Differential effects of HNF-1α mutations associated with familial young-onset diabetes on target gene regulation
- PMID: 21170474
- PMCID: PMC3060974
- DOI: 10.2119/molmed.2010.00097
Differential effects of HNF-1α mutations associated with familial young-onset diabetes on target gene regulation
Abstract
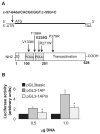
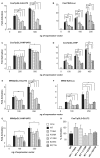
Hepatocyte nuclear factor 1-α (HNF-1α) is a homeodomain transcription factor expressed in a variety of tissues (including liver and pancreas) that regulates a wide range of genes. Heterozygous mutations in the gene encoding HNF-1α (HNF1A) cause familial young-onset diabetes, also known as maturity-onset diabetes of the young, type 3 (MODY3). The variability of the MODY3 clinical phenotype can be due to environmental and genetic factors as well as to the type and position of mutations. Thus, functional characterization of HNF1A mutations might provide insight into the molecular defects explaining the variability of the MODY3 phenotype. We have functionally characterized six HNF1A mutations identified in diabetic patients: two novel ones, p.Glu235Gly and c-57-64delCACGCGGT;c-55G>C; and four previously described, p.Val133Met, p.Thr196Ala, p.Arg271Trp and p.Pro379Arg. The effects of mutations on transcriptional activity have been measured by reporter assays on a subset of HNF-1α target promoters in Cos7 and Min6 cells. Target DNA binding affinities have been quantified by electrophoretic mobility shift assay using bacterially expressed glutathione-S-transferase (GST)-HNF-1α fusion proteins and nuclear extracts of transfected Cos7 cells. Our functional studies revealed that mutation c-57-64delCACGCGGT;c-55G>C reduces HNF1A promoter activity in Min6 cells and that missense mutations have variable effects. Mutation p.Arg271Trp impairs HNF-1α activity in all conditions tested, whereas mutations p.Val133Met, p.Glu235Gly and p.Pro379Arg exert differential effects depending on the target promoter. In contrast, substitution p.Thr196Ala does not appear to alter HNF-1α function. Our results suggest that HNF1A mutations may have differential effects on the regulation of specific target genes, which could contribute to the variability of the MODY3 clinical phenotype.
Figures


References
-
- Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008;4:200–13. - PubMed
-
- Mendel DB, Crabtree GR. HNF-1, a member of a novel class of dimerizing homeodomain proteins. J Biol Chem. 1991;266:677–80. - PubMed
-
- Shih DQ, et al. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001;50:2472–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

