Bottom-up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology
- PMID: 21171616
- PMCID: PMC3010238
- DOI: 10.1021/nl0494497
Bottom-up Assembly of RNA Arrays and Superstructures as Potential Parts in Nanotechnology
Abstract
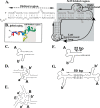
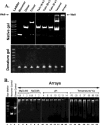
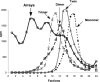
DNA and protein have been extensively scrutinized for feasibility as parts in nanotechnology, but another natural building block, RNA, has been largely ignored. RNA can be manipulated to form versatile shapes, thus providing an element of adaptability to DNA nanotechnology, which is predominantly based upon a double-helical structure. The DNA-packaging motor of bacterial virus phi29 contains six DNA-packaging RNAs (pRNA), which together form a hexameric ring via loop/loop interaction. Here we report that this pRNA can be redesigned to form a variety of structures and shapes, including twins, tetramers, rods, triangles, and 3D arrays several microns in size via interaction of programmed helical regions and loops. Three dimensional RNA array formation required a defined nucleotide number for twisting of the interactive helix and a palindromic sequence. Such arrays are unusually stable and resistant to a wide range of temperatures, salt concentrations, and pH.
Figures


References
-
- Mao C, LaBean TH, Relf JH, Seeman NC. Nature. 2000;407:493–496. - PubMed
-
- Williams KA, Veenhuizen PT, de la Torre BG, Eritja R, Dekker C. Nature. 2002;420(6917):761. - PubMed
-
- Mao C, Solis DJ, Reiss BD, Kottmann ST, Sweeney RY, Hayhurst A, Georgiou G, Iverson B, Belcher AM. Science. 2004;303:213–217. - PubMed
-
- Soong RK, Bachand GD, Neves HP, Olkhovets AG, Craighead HG, Montemagno CD. Science. 2000;290:1555–1558. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
