An endogenous murine leukemia viral genome contaminant in a commercial RT-PCR kit is amplified using standard primers for XMRV
- PMID: 21171978
- PMCID: PMC3024226
- DOI: 10.1186/1742-4690-7-110
An endogenous murine leukemia viral genome contaminant in a commercial RT-PCR kit is amplified using standard primers for XMRV
Abstract
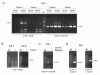
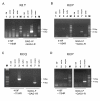
During pilot studies to investigate the presence of viral RNA of xenotropic murine leukemia virus (MLV)-related virus (XMRV) infection in sera from chronic fatigue syndrome (CFS) patients in Japan, a positive band was frequently detected at the expected product size in negative control samples when detecting a partial gag region of XMRV using a one-step RT-PCR kit. We suspected that the kit itself might have been contaminated with small traces of endogenous MLV genome or XMRV and attempted to evaluate the quality of the kit in two independent laboratories. We purchased four one-step RT-PCR kits from Invitrogen, TaKaRa, Promega and QIAGEN in Japan. To amplify the partial gag gene of XMRV or other MLV-related viruses, primer sets (419F and 1154R, and GAG-I-F and GAG-I-R) which have been widely used in XMRV studies were employed. The nucleotide sequences of the amplicons were determined and compared with deposited sequences of a polytropic endogenous MLV (PmERV), XMRV and endogenous MLV-related viruses derived from CFS patients. We found that the enzyme mixtures of the one-step RT-PCR kit from Invitrogen were contaminated with RNA derived from PmERV. The nucleotide sequence of a partial gag region of the contaminant amplified by RT-PCR was nearly identical (99.4% identity) to a PmERV on chromosome 7 and highly similar (96.9 to 97.6%) to recently identified MLV-like viruses derived from CFS patients. We also determined the nucleotide sequence of a partial env region of the contaminant and found that it was almost identical (99.6%) to the PmERV. In the investigation of XMRV infection in patients of CFS and prostate cancer, researchers should prudently evaluate the test kits for the presence of endogenous MLV as well as XMRV genomes prior to PCR and RT-PCR tests.
Figures


References
-
- Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, DeRisi JL. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. - DOI - PMC - PubMed
-
- Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Silverman RH, Mikovits JA. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:585–589. doi: 10.1126/science.1179052. - DOI - PubMed
-
- Groom HC, Boucherit VC, Makinson K, Randal E, Baptista S, Hagan S, Gow JW, Mattes FM, Breuer J, Kerr JR, Stoye JP, Bishop KN. Absence of xenotropic murine leukaemia virus-related virus in UK patients with chronic fatigue syndrome. Retrovirology. 2010;7:10. doi: 10.1186/1742-4690-7-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

