Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)?
- PMID: 21172831
- PMCID: PMC3107663
- DOI: 10.1093/molbev/msq345
Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)?
Abstract
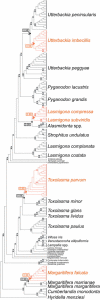
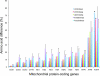
Mitochondrial (mt) function depends critically on optimal interactions between components encoded by mt and nuclear DNAs. mitochondrial DNA (mtDNA) inheritance (SMI) is thought to have evolved in animal species to maintain mito-nuclear complementarity by preventing the spread of selfish mt elements thus typically rendering mtDNA heteroplasmy evolutionarily ephemeral. Here, we show that mtDNA intraorganismal heteroplasmy can have deterministic underpinnings and persist for hundreds of millions of years. We demonstrate that the only exception to SMI in the animal kingdom, that is, the doubly uniparental mtDNA inheritance system in bivalves, with its three-way interactions among egg mt-, sperm mt- and nucleus-encoded gene products, is tightly associated with the maintenance of separate male and female sexes (dioecy) in freshwater mussels. Specifically, this mother-through-daughter and father-through-son mtDNA inheritance system, containing highly differentiated mt genomes, is found in all dioecious freshwater mussel species. Conversely, all hermaphroditic species lack the paternally transmitted mtDNA (=possess SMI) and have heterogeneous macromutations in the recently discovered, novel protein-coding gene (F-orf) in their maternally transmitted mt genomes. Using immunoelectron microscopy, we have localized the F-open reading frame (ORF) protein, likely involved in specifying separate sexes, in mitochondria and in the nucleus. Our results support the hypothesis that proteins coded by the highly divergent maternally and paternally transmitted mt genomes could be directly involved in sex determination in freshwater mussels. Concomitantly, our study demonstrates novel features for animal mt genomes: the existence of additional, lineage-specific, mtDNA-encoded proteins with functional significance and the involvement of mtDNA-encoded proteins in extra-mt functions. Our results open new avenues for the identification, characterization, and functional analyses of ORFs in the intergenic regions, previously defined as "noncoding," found in a large proportion of animal mt genomes.
Figures



References
-
- Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

