Selective ploidy ablation, a high-throughput plasmid transfer protocol, identifies new genes affecting topoisomerase I-induced DNA damage
- PMID: 21173034
- PMCID: PMC3044861
- DOI: 10.1101/gr.109033.110
Selective ploidy ablation, a high-throughput plasmid transfer protocol, identifies new genes affecting topoisomerase I-induced DNA damage
Abstract
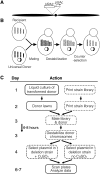
We have streamlined the process of transferring plasmids into any yeast strain library by developing a novel mating-based, high-throughput method called selective ploidy ablation (SPA). SPA uses a universal plasmid donor strain that contains conditional centromeres on every chromosome. The plasmid-bearing donor is mated to a recipient, followed by removal of all donor-strain chromosomes, producing a haploid strain containing the transferred plasmid. As proof of principle, we used SPA to transfer plasmids containing wild-type and mutant alleles of DNA topoisomerase I (TOP1) into the haploid yeast gene-disruption library. Overexpression of Top1 identified only one sensitive mutation, rpa34, while overexpression of top1-T(722)A allele, a camptothecin mimetic, identified 190 sensitive gene-disruption strains along with rpa34. In addition to known camptothecin-sensitive strains, this set contained mutations in genes involved in the Rpd3 histone deacetylase complex, the kinetochore, and vesicle trafficking. We further show that mutations in several ESCRT vesicle trafficking components increase Top1 levels, which is dependent on SUMO modification. These findings demonstrate the utility of the SPA technique to introduce plasmids into the haploid gene-disruption library to discover new interacting pathways.
Figures


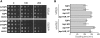

References
-
- Alvaro D, Sunjevaric I, Reid RJ, Lisby M, Stillman DJ, Rothstein R 2006. Systematic hybrid LOH: A new method to reduce false positives and negatives during screening of yeast gene deletion libraries. Yeast 23: 1097–1106 - PubMed
-
- Branzei D, Foiani M 2010. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
