Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin
- PMID: 21173115
- PMCID: PMC3010068
- DOI: 10.1083/jcb.201007013
Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin
Abstract
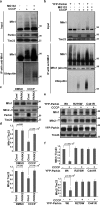
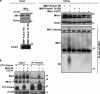
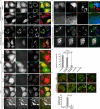
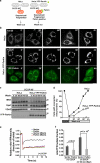
Damage to mitochondria can lead to the depolarization of the inner mitochondrial membrane, thereby sensitizing impaired mitochondria for selective elimination by autophagy. However, fusion of uncoupled mitochondria with polarized mitochondria can compensate for damage, reverse membrane depolarization, and obviate mitophagy. Parkin, an E3 ubiquitin ligase that is mutated in monogenic forms of Parkinson's disease, was recently found to induce selective autophagy of damaged mitochondria. Here we show that ubiquitination of mitofusins Mfn1 and Mfn2, large GTPases that mediate mitochondrial fusion, is induced by Parkin upon membrane depolarization and leads to their degradation in a proteasome- and p97-dependent manner. p97, a AAA+ ATPase, accumulates on mitochondria upon uncoupling of Parkin-expressing cells, and both p97 and proteasome activity are required for Parkin-mediated mitophagy. After mitochondrial fission upon depolarization, Parkin prevents or delays refusion of mitochondria, likely by the elimination of mitofusins. Inhibition of Drp1-mediated mitochondrial fission, the proteasome, or p97 prevents Parkin-induced mitophagy.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

