Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories
- PMID: 21173116
- PMCID: PMC3010067
- DOI: 10.1083/jcb.201007074
Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories
Abstract
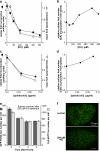
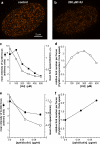
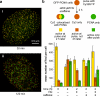
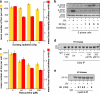
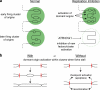
Replication origins are licensed by loading MCM2-7 hexamers before entry into S phase. However, only ∼10% of licensed origins are normally used in S phase, with the others remaining dormant. When fork progression is inhibited, dormant origins initiate nearby to ensure that all of the DNA is eventually replicated. In apparent contrast, replicative stress activates ataxia telangiectasia and rad-3-related (ATR) and Chk1 checkpoint kinases that inhibit origin firing. In this study, we show that at low levels of replication stress, ATR/Chk1 predominantly suppresses origin initiation by inhibiting the activation of new replication factories, thereby reducing the number of active factories. At the same time, inhibition of replication fork progression allows dormant origins to initiate within existing replication factories. The inhibition of new factory activation by ATR/Chk1 therefore redirects replication toward active factories where forks are inhibited and away from regions that have yet to start replication. This minimizes the deleterious consequences of fork stalling and prevents similar problems from arising in unreplicated regions of the genome.
Figures


Similar articles
-
Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus.J Cell Sci. 2004 Dec 1;117(Pt 25):6019-30. doi: 10.1242/jcs.01400. Epub 2004 Nov 9. J Cell Sci. 2004. PMID: 15536124 Free PMC article.
-
Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage.Mol Cell Biol. 2005 May;25(9):3553-62. doi: 10.1128/MCB.25.9.3553-3562.2005. Mol Cell Biol. 2005. PMID: 15831461 Free PMC article.
-
Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing.J Cell Biol. 2001 Sep 3;154(5):913-23. doi: 10.1083/jcb.200104099. J Cell Biol. 2001. PMID: 11535615 Free PMC article.
-
Dormant origin signaling during unperturbed replication.DNA Repair (Amst). 2019 Sep;81:102655. doi: 10.1016/j.dnarep.2019.102655. Epub 2019 Jul 8. DNA Repair (Amst). 2019. PMID: 31311769 Free PMC article. Review.
-
Dormant origins, the licensing checkpoint, and the response to replicative stresses.Cold Spring Harb Perspect Biol. 2012 Oct 1;4(10):a012955. doi: 10.1101/cshperspect.a012955. Cold Spring Harb Perspect Biol. 2012. PMID: 22904560 Free PMC article. Review.
Cited by
-
Crosslinks and replication: the show must go on.Cell Cycle. 2015;14(17):2721-2. doi: 10.1080/15384101.2015.1010971. Epub 2015 Feb 26. Cell Cycle. 2015. PMID: 25719851 Free PMC article. No abstract available.
-
Accurate delineation of cell cycle phase transitions in living cells with PIP-FUCCI.Cell Cycle. 2018;17(21-22):2496-2516. doi: 10.1080/15384101.2018.1547001. Cell Cycle. 2018. PMID: 30421640 Free PMC article.
-
Genome maintenance meets mechanobiology.Chromosoma. 2024 Jan;133(1):15-36. doi: 10.1007/s00412-023-00807-5. Epub 2023 Aug 15. Chromosoma. 2024. PMID: 37581649 Free PMC article. Review.
-
On the Interplay of the DNA Replication Program and the Intra-S Phase Checkpoint Pathway.Genes (Basel). 2019 Jan 29;10(2):94. doi: 10.3390/genes10020094. Genes (Basel). 2019. PMID: 30700024 Free PMC article. Review.
-
Combined use of subclinical hydroxyurea and CHK1 inhibitor effectively controls melanoma and lung cancer progression, with reduced normal tissue toxicity compared to gemcitabine.Mol Oncol. 2019 Jul;13(7):1503-1518. doi: 10.1002/1878-0261.12497. Epub 2019 Jun 14. Mol Oncol. 2019. PMID: 31044505 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

