Sphingosine-1-phosphate lyase deficiency produces a pro-inflammatory response while impairing neutrophil trafficking
- PMID: 21173151
- PMCID: PMC3044991
- DOI: 10.1074/jbc.M110.171819
Sphingosine-1-phosphate lyase deficiency produces a pro-inflammatory response while impairing neutrophil trafficking
Abstract
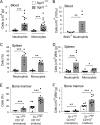
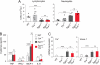
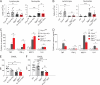
Sphingosine-1-phosphate (S1P) lyase catalyzes the degradation of S1P, a potent signaling lysosphingolipid. Mice with an inactive S1P lyase gene are impaired in the capacity to degrade S1P, resulting in highly elevated S1P levels. These S1P lyase-deficient mice have low numbers of lymphocytes and high numbers of neutrophils in their blood. We found that the S1P lyase-deficient mice exhibited features of an inflammatory response including elevated levels of pro-inflammatory cytokines and an increased expression of genes in liver associated with an acute-phase response. However, the recruitment of their neutrophils into inflamed tissues was impaired and their neutrophils were defective in migration to chemotactic stimulus. The IL-23/IL-17/granulocyte-colony stimulating factor (G-CSF) cytokine-controlled loop regulating neutrophil homeostasis, which is dependent on neutrophil trafficking to tissues, was disturbed in S1P lyase-deficient mice. Deletion of the S1P4 receptor partially decreased the neutrophilia and inflammation in S1P lyase-deficient mice, implicating S1P receptor signaling in the phenotype. Thus, a genetic block in S1P degradation elicits a pro-inflammatory response but impairs neutrophil migration from blood into tissues.
Figures
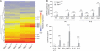




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

