Site-specific mutation of Staphylococcus aureus VraS reveals a crucial role for the VraR-VraS sensor in the emergence of glycopeptide resistance
- PMID: 21173175
- PMCID: PMC3067069
- DOI: 10.1128/AAC.00720-10
Site-specific mutation of Staphylococcus aureus VraS reveals a crucial role for the VraR-VraS sensor in the emergence of glycopeptide resistance
Abstract
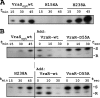
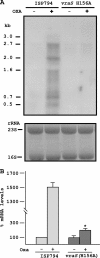
An initial response of Staphylococcus aureus to encounter with cell wall-active antibiotics occurs by transmembrane signaling systems that orchestrate changes in gene expression to promote survival. Histidine kinase two-component sensor-response regulators such as VraRS contribute to this response. In this study, we examined VraS membrane sensor phosphotransfer signal transduction and explored the genetic consequences of disrupting signaling by engineering a site-specific vraS chromosomal mutation. We have used in vitro autophosphorylation assay with purified VraS[64-347] lacking its transmembrane anchor region and tested site-specific kinase domain histidine mutants. We identified VraS H156 as the probable site of autophosphorylation and show phosphotransfer in vitro using purified VraR. Genetic studies show that the vraS(H156A) mutation in three strain backgrounds (ISP794, Newman, and COL) fails to generate detectable first-step reduced susceptibility teicoplanin mutants and severely reduces first-step vancomycin mutants. The emergence of low-level glycopeptide resistance in strain ISP794, derived from strain 8325 (ΔrsbU), did not require a functional σ(B), but rsbU restoration could enhance the emergence frequency supporting a role for this alternative sigma factor in promoting glycopeptide resistance. Transcriptional analysis of vraS(H156A) strains revealed a pronounced reduction but not complete abrogation of the vraRS operon after exposure to cell wall-active antibiotics, suggesting that additional factors independent of VraS-driven phosphotransfer, or σ(B), exist for this promoter. Collectively, our results reveal important details of the VraRS signaling system and predict that pharmacologic blockade of the VraS sensor kinase will have profound effects on blocking emergence of cell wall-active antibiotic resistance in S. aureus.
Figures


Similar articles
-
NH125 Sensitizes Staphylococcus aureus to Cell Wall-Targeting Antibiotics through the Inhibition of the VraS Sensor Histidine Kinase.Microbiol Spectr. 2023 Jun 15;11(3):e0486122. doi: 10.1128/spectrum.04861-22. Epub 2023 May 25. Microbiol Spectr. 2023. PMID: 37227302 Free PMC article.
-
Genetic changes associated with glycopeptide resistance in Staphylococcus aureus: predominance of amino acid substitutions in YvqF/VraSR.J Antimicrob Chemother. 2010 Jan;65(1):37-45. doi: 10.1093/jac/dkp394. J Antimicrob Chemother. 2010. PMID: 19889788 Free PMC article.
-
The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus.FEMS Microbiol Lett. 2006 Sep;262(2):163-71. doi: 10.1111/j.1574-6968.2006.00384.x. FEMS Microbiol Lett. 2006. PMID: 16923071
-
Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus.Antimicrob Agents Chemother. 2011 Apr;55(4):1391-402. doi: 10.1128/AAC.01213-10. Epub 2011 Jan 10. Antimicrob Agents Chemother. 2011. PMID: 21220524 Free PMC article.
-
Glycopeptide resistance in Staphylococcus aureus: is it a real threat?J Infect Chemother. 2004 Apr;10(2):69-75. doi: 10.1007/s10156-004-0307-5. J Infect Chemother. 2004. PMID: 15160298 Review.
Cited by
-
Synergistic use of anti-inflammatory ketorolac and gentamicin to target staphylococcal biofilms.Res Sq [Preprint]. 2023 Oct 26:rs.3.rs-3471646. doi: 10.21203/rs.3.rs-3471646/v1. Res Sq. 2023. Update in: J Transl Med. 2024 Jan 25;22(1):102. doi: 10.1186/s12967-024-04871-y. PMID: 37961705 Free PMC article. Updated. Preprint.
-
Combatting Antibiotic-Resistant Staphylococcus aureus: Discovery of TST1N-224, a Potent Inhibitor Targeting Response Regulator VraRC, through Pharmacophore-Based Screening and Molecular Characterizations.J Chem Inf Model. 2024 Aug 12;64(15):6132-6146. doi: 10.1021/acs.jcim.4c01046. Epub 2024 Jul 30. J Chem Inf Model. 2024. PMID: 39078379 Free PMC article.
-
Revisiting the Role of VraTSR in Staphylococcus aureus Response to Cell Wall-Targeting Antibiotics.J Bacteriol. 2022 Aug 16;204(8):e0016222. doi: 10.1128/jb.00162-22. Epub 2022 Jul 13. J Bacteriol. 2022. PMID: 35862765 Free PMC article.
-
NH125 Sensitizes Staphylococcus aureus to Cell Wall-Targeting Antibiotics through the Inhibition of the VraS Sensor Histidine Kinase.Microbiol Spectr. 2023 Jun 15;11(3):e0486122. doi: 10.1128/spectrum.04861-22. Epub 2023 May 25. Microbiol Spectr. 2023. PMID: 37227302 Free PMC article.
-
Evolution under vancomycin selection drives divergent collateral sensitivity patterns in Staphylococcus aureus.bioRxiv [Preprint]. 2025 Apr 5:2023.11.30.569373. doi: 10.1101/2023.11.30.569373. bioRxiv. 2025. PMID: 38077036 Free PMC article. Preprint.
References
-
- Belcheva, A., and D. Golemi-Kotra. 2008. A close-up view of the VraSR two-component system: a mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 283:12354-12364. - PubMed
-
- Belcheva, A., V. Verma, and D. Golemi-Kotra. 2009. DNA-binding activity of the vancomycin resistance associated regulator protein VraR and the role of phosphorylation in transcriptional regulation of the vraSR operon. Biochemistry 48:5592-5601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

