Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells
- PMID: 21173232
- PMCID: PMC3017192
- DOI: 10.1073/pnas.1013004108
Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells
Abstract
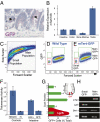
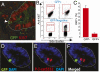
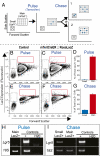
The intestinal epithelium is maintained by a population of rapidly cycling (Lgr5(+)) intestinal stem cells (ISCs). It has been postulated, however, that slowly cycling ISCs must also be present in the intestine to protect the genome from accumulating deleterious mutations and to allow for a response to tissue injury. Here, we identify a subpopulation of slowly cycling ISCs marked by mouse telomerase reverse transcriptase (mTert) expression that can give rise to Lgr5(+) cells. mTert-expressing cells distribute in a pattern along the crypt-villus axis similar to long-term label-retaining cells (LRCs) and are resistant to tissue injury. Lineage-tracing studies demonstrate that mTert(+) cells give rise to all differentiated intestinal cell types, persist long term, and contribute to the regenerative response following injury. Consistent with other highly regenerative tissues, our results demonstrate that a slowly cycling stem cell population exists within the intestine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

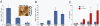
References
-
- Potten CS, Loeffler M. Stem cells: Attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. - PubMed
-
- Scoville DH, Sato T, He XC, Li L. Current view: Intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. - PubMed
-
- Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- van der Flier LG, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK 084056/DK/NIDDK NIH HHS/United States
- R37 DK 32658/DK/NIDDK NIH HHS/United States
- R01 DK084056/DK/NIDDK NIH HHS/United States
- P30 HD 18655/HD/NICHD NIH HHS/United States
- K08 DK066305/DK/NIDDK NIH HHS/United States
- K08 DK 066305/DK/NIDDK NIH HHS/United States
- R21 DK 078198/DK/NIDDK NIH HHS/United States
- R37 DK032658/DK/NIDDK NIH HHS/United States
- P30 HD018655/HD/NICHD NIH HHS/United States
- P30 DK034854/DK/NIDDK NIH HHS/United States
- P30 DK 34854/DK/NIDDK NIH HHS/United States
- R21 DK078198/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

