Effective population size and the efficacy of selection on the X chromosomes of two closely related Drosophila species
- PMID: 21173424
- PMCID: PMC3038356
- DOI: 10.1093/gbe/evq086
Effective population size and the efficacy of selection on the X chromosomes of two closely related Drosophila species
Abstract
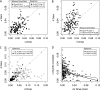
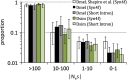
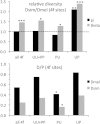
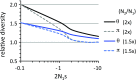
The prevalence of natural selection relative to genetic drift is of central interest in evolutionary biology. Depending on the distribution of fitness effects of new mutations, the importance of these evolutionary forces may differ in species with different effective population sizes. Here, we survey population genetic variation at 105 orthologous X-linked protein coding regions in Drosophila melanogaster and its sister species D. simulans, two closely related species with distinct demographic histories. We observe significantly higher levels of polymorphism and evidence for stronger selection on codon usage bias in D. simulans, consistent with a larger historical effective population size on average for this species. Despite these differences, we estimate that <10% of newly arising nonsynonymous mutations have deleterious fitness effects in the nearly neutral range (i.e., -10 < N(e)s < 0) in both species. The inferred distributions of fitness effects and demographic models translate into surprisingly high estimates of the fraction of "adaptive" protein divergence in both species (∼ 85-90%). Despite evidence for different demographic histories, differences in population size have apparently played little role in the dynamics of protein evolution in these two species, and estimates of the adaptive fraction (α) of protein divergence in both species remain high even if we account for recent 10-fold growth. Furthermore, although several recent studies have noted strong signatures of recurrent adaptive protein evolution at genes involved in immunity, reproduction, sexual conflict, and intragenomic conflict, our finding of high levels of adaptive protein divergence at randomly chosen proteins (with respect to function) suggests that many other factors likely contribute to the adaptive protein divergence signature in Drosophila.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

