Fetal environment, epigenetics, and pediatric renal disease
- PMID: 21174217
- PMCID: PMC3063864
- DOI: 10.1007/s00467-010-1714-8
Fetal environment, epigenetics, and pediatric renal disease
Abstract
The notion that some adult diseases may have their origins in utero has recently captured scientists' attention. Some of these effects persist across generations and may involve epigenetic mechanisms. Epigenetic modifications, DNA methylation together with covalent modifications of histones, alter chromatin density and accessibility of DNA to cellular machinery, modulating the transcriptional potential of the underlying DNA sequence. Here, we will discuss the different epigenetic modifications and their potential role in and contribution to renal disease development.
Figures

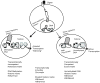
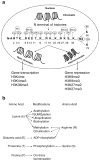
References
-
- Knoll JH, Nicholls RD, Magenis RE, Graham JM, Jr, Lalande M, Latt SA. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet. 1989;32:285–290. - PubMed
-
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

