NFKBIA deletion in glioblastomas
- PMID: 21175304
- PMCID: PMC3652611
- DOI: 10.1056/NEJMoa1006312
NFKBIA deletion in glioblastomas
Abstract
Background: Amplification and activating mutations of the epidermal growth factor receptor (EGFR) oncogene are molecular hallmarks of glioblastomas. We hypothesized that deletion of NFKBIA (encoding nuclear factor of κ-light polypeptide gene enhancer in B-cells inhibitor-α), an inhibitor of the EGFR-signaling pathway, promotes tumorigenesis in glioblastomas that do not have alterations of EGFR.
Methods: We analyzed 790 human glioblastomas for deletions, mutations, or expression of NFKBIA and EGFR. We studied the tumor-suppressor activity of NFKBIA in tumor-cell culture. We compared the molecular results with the outcome of glioblastoma in 570 affected persons.
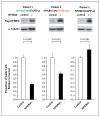
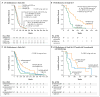
Results: NFKBIA is often deleted but not mutated in glioblastomas; most deletions occur in nonclassical subtypes of the disease. Deletion of NFKBIA and amplification of EGFR show a pattern of mutual exclusivity. Restoration of the expression of NFKBIA attenuated the malignant phenotype and increased the vulnerability to chemotherapy of cells cultured from tumors with NFKBIA deletion; it also reduced the viability of cells with EGFR amplification but not of cells with normal gene dosages of both NFKBIA and EGFR. Deletion and low expression of NFKBIA were associated with unfavorable outcomes. Patients who had tumors with NFKBIA deletion had outcomes that were similar to those in patients with tumors harboring EGFR amplification. These outcomes were poor as compared with the outcomes in patients with tumors that had normal gene dosages of NFKBIA and EGFR. A two-gene model that was based on expression of NFKBIA and O(6)-methylguanine DNA methyltransferase was strongly associated with the clinical course of the disease.
Conclusions: Deletion of NFKBIA has an effect that is similar to the effect of EGFR amplification in the pathogenesis of glioblastoma and is associated with comparatively short survival.
Figures


Comment in
-
Monoallelic deletion of NFKBIA in glioblastoma: when less is more.Cancer Cell. 2011 Feb 15;19(2):163-5. doi: 10.1016/j.ccr.2011.01.045. Cancer Cell. 2011. PMID: 21316598
-
NFKBIA deletion in glioblastomas.N Engl J Med. 2011 Jul 21;365(3):277; author reply 277-8. doi: 10.1056/NEJMc1103354. N Engl J Med. 2011. PMID: 21774727 No abstract available.
-
NFKBIA deletion in glioblastomas.N Engl J Med. 2011 Jul 21;365(3):276-7; author reply 277-8. doi: 10.1056/NEJMc1103354. N Engl J Med. 2011. PMID: 21774728 No abstract available.
References
-
- Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. [Erratum, N Engl J Med 2008;359:877.] - PubMed
-
- Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
