Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3'-termini
- PMID: 21175352
- PMCID: PMC3518376
- DOI: 10.1667/RR2332.1
Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3'-termini
Abstract
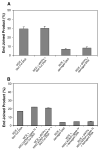
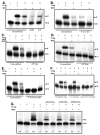
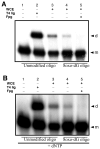
Radiation-induced DNA double-strand breaks (DSBs) are critical cytotoxic lesions that are typically repaired by nonhomologous end joining (NHEJ) in human cells. Our previous work indicated that the highly cytotoxic DSBs formed by (125)I decay possess base damage clustered within 8 to 10 bases of the break and 3'-phosphate (P) and 3'-OH ends. This study examined the effect of such structures on NHEJ in in vitro assays employing either (125)I decay-induced DSB linearized plasmid DNA or structurally defined duplex oligonucleotides. Duplex oligonucleotides that possess either a 3'-P or 3'-phosphoglycolate (PG) or a ligatable 3'-OH end with either an AP site or an 8-oxo-dG 1 nucleotide upstream (-1n) from the 3'-terminus have been examined for reparability. Moderate to severe end-joining inhibition was observed for modified DSB ends or 8-oxo-dG upstream from a 3'-OH end. In contrast, abolition of end joining was observed with duplexes possessing an AP site upstream from a ligatable 3'-OH end or for a lesion combination involving 3'-P plus an upstream 8-oxo-dG. In addition, base mismatches at the -1n position were also strong inhibitors of NHEJ in this system, suggesting that destabilization of the DSB terminus as a result of base loss or improper base pairing may play a role in the inhibitory effects of these structures. Furthermore, we provide data indicating that DSB end joining is likely to occur prior to removal or repair of base lesions proximal to the DSB terminus. Our results show that base damage or base loss near a DSB end may be a severe block to NHEJ and that complex combinations of lesions presented in the context of a DSB may be more inhibitory than the individual lesions alone. In contrast, blocked DSB 3'-ends alone are only modestly inhibitory to NHEJ. Finally, DNA ligase activity is implicated as being responsible for these effects.
Figures



References
-
- Ward JF. Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat Res. 1981;86:185–195. - PubMed
-
- Iliakis G. The role of DNA double strand breaks in ionizing radiation-induced killing of eukaryotic cells. Bioessays. 1991;13:641–648. - PubMed
-
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. - PubMed
-
- Pastwa E, Neumann RD, Mezhevaya K, Winters TA. Repair of radiation-induced DNA double-strand breaks is dependent upon radiation quality and the structural complexity of double-strand breaks. Radiat Res. 2003;159:251–261. - PubMed
-
- Radford IR. DNA lesion complexity and induction of apoptosis by ionizing radiation. Int J Radiat Biol. 2002;78:457–466. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

