Tyrosine hydroxylase and regulation of dopamine synthesis
- PMID: 21176768
- PMCID: PMC3065393
- DOI: 10.1016/j.abb.2010.12.017
Tyrosine hydroxylase and regulation of dopamine synthesis
Abstract
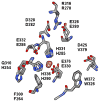
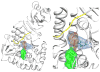
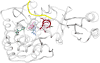
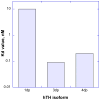
Tyrosine hydroxylase is the rate-limiting enzyme of catecholamine biosynthesis; it uses tetrahydrobiopterin and molecular oxygen to convert tyrosine to DOPA. Its amino terminal 150 amino acids comprise a domain whose structure is involved in regulating the enzyme's activity. Modes of regulation include phosphorylation by multiple kinases at four different serine residues, and dephosphorylation by two phosphatases. The enzyme is inhibited in feedback fashion by the catecholamine neurotransmitters. Dopamine binds to TyrH competitively with tetrahydrobiopterin, and interacts with the R domain. TyrH activity is modulated by protein-protein interactions with enzymes in the same pathway or the tetrahydrobiopterin pathway, structural proteins considered to be chaperones that mediate the neuron's oxidative state, and the protein that transfers dopamine into secretory vesicles. TyrH is modified in the presence of NO, resulting in nitration of tyrosine residues and the glutathionylation of cysteine residues.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures









References
-
- Molinoff PB, Axelrod J. Biochemistry of catecholamines. Annual Review of Biochemistry. 1971;40:465–500. - PubMed
-
- Weiner N. Tyrosine-3-monooxygenase (tyrosine hydroxylase) In: Youdim MBH, editor. Aromatic amino acid hydroxylases and mental disease. John Wiley & Sons, Ltd; New York: 1979. pp. 141–190.
-
- Tripp G, Wickens JR. Neurobiology of adhd. Neuropharmacology. 2009;57:579–589. - PubMed
-
- Arnsten AFT. Catecholamine regulation of the prefrontal cortex. J Psychopharmacol. 1997;11:151–162. - PubMed
-
- Assadi SM, Yücel M, Pantelis C. Dopamine modulates neural networks involved in effort-based decision-making. Neuroscience & Biobehavioral Reviews. 2009;33:383–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

