Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity
- PMID: 21177245
- PMCID: PMC3044941
- DOI: 10.1074/jbc.R110.209502
Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity
Abstract
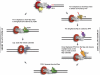
In eukaryotic nuclear DNA replication, one strand of DNA is synthesized continuously, but the other is made as Okazaki fragments that are later joined. Discontinuous synthesis is inherently more complex, and fragmented intermediates create risks for disruptions of genome integrity. Genetic analyses and biochemical reconstitutions indicate that several parallel pathways evolved to ensure that the fragments are made and joined with integrity. An RNA primer is removed from each fragment before joining by a process involving polymerase-dependent displacement into a single-stranded flap. Evidence in vitro suggests that, with most fragments, short flaps are displaced and efficiently cleaved. Some flaps can become long, but these are also removed to allow joining. Rarely, a flap can form structure, necessitating displacement of the entire fragment. There is now evidence that post-translational protein modification regulates the flow through the pathways to favor protection of genomic information in regions of actively transcribed chromatin.
Figures


References
-
- Kornberg A., Baker T. A. (1992) DNA Replication, 2nd Ed., W. H. Freeman, New York
-
- Sakabe K., Okazaki R. (1966) Biochim. Biophys. Acta 129, 651–654 - PubMed
-
- Alberts B. M., Barry J., Bedinger P., Formosa T., Jongeneel C. V., Kreuzer K. N. (1983) Cold Spring Harbor Symp. Quant. Biol. 47, 655–668 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

