Differential G-protein-coupled receptor phosphorylation provides evidence for a signaling bar code
- PMID: 21177246
- PMCID: PMC3064205
- DOI: 10.1074/jbc.M110.154526
Differential G-protein-coupled receptor phosphorylation provides evidence for a signaling bar code
Abstract
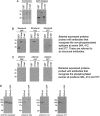
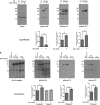
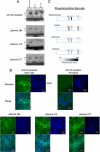
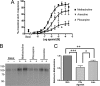
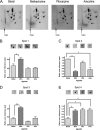
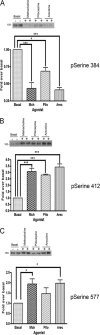
G-protein-coupled receptors are hyper-phosphorylated in a process that controls receptor coupling to downstream signaling pathways. The pattern of receptor phosphorylation has been proposed to generate a "bar code" that can be varied in a tissue-specific manner to direct physiologically relevant receptor signaling. If such a mechanism existed, receptors would be expected to be phosphorylated in a cell/tissue-specific manner. Using tryptic phosphopeptide maps, mass spectrometry, and phospho-specific antibodies, it was determined here that the prototypical G(q/11)-coupled M(3)-muscarinic receptor was indeed differentially phosphorylated in various cell and tissue types supporting a role for differential receptor phosphorylation in directing tissue-specific signaling. Furthermore, the phosphorylation profile of the M(3)-muscarinic receptor was also dependent on the stimulus. Full and partial agonists to the M(3)-muscarinic receptor were observed to direct phosphorylation preferentially to specific sites. This hitherto unappreciated property of ligands raises the possibility that one mechanism underlying ligand bias/functional selectivity, a process where ligands direct receptors to preferred signaling pathways, may be centered on the capacity of ligands to promote receptor phosphorylation at specific sites.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

