Germline epigenetic regulation of KILLIN in Cowden and Cowden-like syndrome
- PMID: 21177507
- PMCID: PMC3655326
- DOI: 10.1001/jama.2010.1877
Germline epigenetic regulation of KILLIN in Cowden and Cowden-like syndrome
Abstract
Context: Germline loss-of-function phosphatase and tensin homolog gene (PTEN) mutations cause 80% of Cowden syndrome, a rare autosomal-dominant disorder (1 in 200,000 live births), characterized by high risks of breast, thyroid, and other cancers. A large heterogeneous group of individuals with Cowden-like syndrome, who have various combinations of Cowden syndrome features but who do not meet Cowden syndrome diagnostic criteria, have PTEN mutations less than 10% of the time, making molecular diagnosis, prediction, genetic counseling, and risk management challenging. Other mechanisms of loss of function such as hypermethylation, which should result in underexpression of PTEN or of KILLIN, a novel tumor suppressor transcribed in the opposite direction, may account for the remainder of Cowden syndrome and Cowden-like syndrome.
Objective: To determine whether germline methylation is found in Cowden syndrome or Cowden-like syndrome in individuals lacking germline PTEN mutations.
Design, setting, and participants: Nucleic acids from prospective nested series of 123 patients with Cowden syndrome or Cowden-like syndrome and 50 unaffected individuals without PTEN variants were analyzed for germline methylation and expression of PTEN and KILLIN at the Cleveland Clinic, August 2008-June 2010. Prevalence of component cancers between groups was compared using the Fisher exact test.
Main outcome measures: Frequency of germline methylation in PTEN mutation-negative Cowden syndrome and Cowden syndrome-like individuals. Prevalence of component cancers in methylation-positive and PTEN mutation-positive individuals.
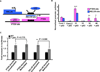
Results: Of 123 patients with Cowden syndrome or Cowden-like syndrome, 45 (37%; 95% confidence interval [CI], 29%-45%) showed hypermethylation upstream of PTEN but no transcriptional repression. The germline methylation was found to transcriptionally down-regulate KILLIN by 250-fold (95% CI, 45-14 286; P = .007) and exclusively disrupted TP53 activation of KILLIN by 30% (95% CI, 7%-45%; P = .008). Demethylation treatment increased only KILLIN expression 4.88-fold (95% CI, 1.4-18.1; P = .05). Individuals with KILLIN -promoter methylation had a 3-fold increased prevalence of breast cancer (35/42 vs 24/64; P < .0001) and a greater than 2-fold increase of kidney cancer (4/45 vs 6/155; P = .004) over individuals with germline PTEN mutations.
Conclusions: Germline KILLIN methylation is common among patients with Cowden syndrome or Cowden-like syndrome and is associated with increased risks of breast and renal cancer over PTEN mutation-positive individuals. These observations need to be replicated.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures
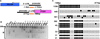


Comment in
-
PTEN promoter silencing and Cowden syndrome: the role of epigenetic regulation of KILLIN.JAMA. 2010 Dec 22;304(24):2744-5. doi: 10.1001/jama.2010.1863. JAMA. 2010. PMID: 21177512 No abstract available.
References
-
- Nelen MR, Padberg GW, Peeters EA, et al. Localization of the gene for Cowden disease to chromosome 10q22-23. Nat Genet. 1996;13:114–116. - PubMed
-
- Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. - PubMed
-
- Marsh DJ, Dahia PL, Zheng Z, et al. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet. 1997;16:333–334. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

