c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation
- PMID: 21177653
- PMCID: PMC3082905
- DOI: 10.1093/nar/gkq1205
c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation
Abstract
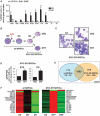
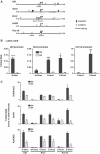
Loss of c-MYC is required for downregulation of ribosomal RNA (rRNA) gene (rDNA) transcription by RNA Polymerase I (Pol I) during granulocyte differentiation. Here, we demonstrate a robust reduction of Pol I loading onto rDNA that along with a depletion of the MYC target gene upstream binding factor (UBF) and a switch from epigenetically active to silent rDNA accompanies this MYC reduction. We hypothesized that MYC may coordinate these mechanisms via direct regulation of multiple components of the Pol I transcription apparatus. Using gene expression arrays we identified a 'regulon' of Pol I factors that are both downregulated during differentiation and reinduced in differentiated granulocytes upon activation of the MYC-ER transgene. This regulon includes the novel c-MYC target genes RRN3 and POLR1B. Although enforced MYC expression during granulocyte differentiation was sufficient to increase the number of active rRNA genes, its activation in terminally differentiated cells did not alter the active to inactive gene ratio despite increased rDNA transcription. Thus, c-MYC dynamically controls rDNA transcription during granulocytic differentiation through the orchestrated transcriptional regulation of core Pol I factors and epigenetic modulation of number of active rRNA genes.
Figures



References
-
- Oskarsson T, Trumpp A. The Myc trilogy: lord of RNA polymerases. Nat. Cell Biol. 2005;7:215–217. - PubMed
-
- Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J, Ramsbottom BA, Innes F, Kantidakis T, Kerr ER, et al. Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochem. Soc. Symp. 2006;73:141–154. - PubMed
-
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr. Biol. 2004;14:R1014–R1027. - PubMed
-
- Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 2003;4:641–649. - PubMed
-
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

