Copy number alterations in urothelial carcinomas: their clinicopathological significance and correlation with DNA methylation alterations
- PMID: 21177765
- PMCID: PMC3066412
- DOI: 10.1093/carcin/bgq274
Copy number alterations in urothelial carcinomas: their clinicopathological significance and correlation with DNA methylation alterations
Abstract
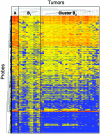
The aim of this study was to clarify the genetic backgrounds underlying the clinicopathological characteristics of urothelial carcinomas (UCs). Array comparative genomic hybridization analysis using a 244K oligonucleotide array was performed on 49 samples of UC tissue. Losses of 2q33.3-q37.3, 4p15.2-q13.1 and 5q13.3-q35.3 and gains of 7p11.2-q11.23 and 20q13.12-q13.2 were correlated with higher histological grade, and gain of 7p21.2-p21.12 was correlated with deeper invasion. Losses of 6q14.1-q27 and 17p13.3-q11.1 and gains of 19q13.12-q13.2 and 20q13.12-q13.33 were correlated with lymph vessel involvement. Loss of 16p12.2-p12.1 and gain of 3q26.32-q29 were correlated with vascular involvement. Losses of 5q14.1-q23.1, 6q14.1-q27, 8p22-p21.3, 11q13.5-q14.1 and 15q11.2-q22.2 and gains of 7p11.2-q11.22 and 19q13.12-q13.2 were correlated with the development of aggressive non-papillary UCs. Losses of 1p32.2-p31.3, 10q11.23-q21.1 and 15q21.3 were correlated with tumor recurrence. Unsupervised hierarchical clustering analysis based on copy number alterations clustered UCs into three subclasses: copy number alterations associated with genome-wide DNA hypomethylation, regional DNA hypermethylation on C-type CpG islands and genome-wide DNA hypo- and hypermethylation were accumulated in clusters A, B(1) and B(2), respectively. Tumor-related genes that may encode therapeutic targets and/or indicators useful for the diagnosis and prognostication of UCs should be explored in the above regions. Both genetic and epigenetic events appear to accumulate during urothelial carcinogenesis, reflecting the clinicopathological diversity of UCs.
Figures


Similar articles
-
Genome-wide DNA methylation profiles in urothelial carcinomas and urothelia at the precancerous stage.Cancer Sci. 2010 Jan;101(1):231-40. doi: 10.1111/j.1349-7006.2009.01330.x. Epub 2009 Aug 27. Cancer Sci. 2010. PMID: 19775289 Free PMC article.
-
Detection of chromosomal alterations in bladder transitional cell carcinomas from Northern China by comparative genomic hybridization.Cancer Lett. 2006 Jul 18;238(2):230-9. doi: 10.1016/j.canlet.2005.07.010. Epub 2005 Aug 25. Cancer Lett. 2006. PMID: 16125302
-
Genetic clustering of clear cell renal cell carcinoma based on array-comparative genomic hybridization: its association with DNA methylation alteration and patient outcome.Clin Cancer Res. 2008 Sep 1;14(17):5531-9. doi: 10.1158/1078-0432.CCR-08-0443. Clin Cancer Res. 2008. PMID: 18765545
-
Investigation of genetic alterations associated with the grade of astrocytic tumor by comparative genomic hybridization.Genes Chromosomes Cancer. 1998 Apr;21(4):340-6. doi: 10.1002/(sici)1098-2264(199804)21:4<340::aid-gcc8>3.0.co;2-z. Genes Chromosomes Cancer. 1998. PMID: 9559346 Review.
-
DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies.Am J Pathol. 1998 May;152(5):1107-23. Am J Pathol. 1998. PMID: 9588877 Free PMC article. Review.
Cited by
-
lncRNA polymorphism affects the prognosis of gastric cancer.World J Surg Oncol. 2022 Aug 31;20(1):273. doi: 10.1186/s12957-022-02723-x. World J Surg Oncol. 2022. PMID: 36045445 Free PMC article.
-
Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity.Nat Rev Cancer. 2015 Jan;15(1):25-41. doi: 10.1038/nrc3817. Nat Rev Cancer. 2015. PMID: 25533674 Review.
-
Hypermethylation in bladder cancer: biological pathways and translational applications.Tumour Biol. 2012 Apr;33(2):347-61. doi: 10.1007/s13277-011-0310-2. Epub 2012 Jan 25. Tumour Biol. 2012. PMID: 22274923
-
Molecular pathological approach to cancer epigenomics and its clinical application.Pathol Int. 2024 Apr;74(4):167-186. doi: 10.1111/pin.13418. Epub 2024 Mar 14. Pathol Int. 2024. PMID: 38482965 Free PMC article. Review.
-
Activation of PPARγ in bladder cancer via introduction of the long arm of human chromosome 9.Oncol Lett. 2022 Mar;23(3):92. doi: 10.3892/ol.2022.13212. Epub 2022 Jan 27. Oncol Lett. 2022. PMID: 35154423 Free PMC article.
References
-
- Kakizoe T, et al. Relationship between papillary and nodular transitional cell carcinoma in the human urinary bladder. Cancer Res. 1988;48:2299–2303. - PubMed
-
- Knowles M, et al. Allelotype of human bladder cancer. Cancer Res. 1994;54:531–538. - PubMed
-
- Orlow I, et al. Deletion of the p16 and p15 genes in human bladder tumors. J. Natl Cancer Inst. 1995;87:1524–1529. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

