Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition
- PMID: 21177896
- PMCID: PMC3067754
- DOI: 10.1128/JCM.01507-10
Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition
Abstract
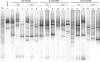
Clostridium difficile is a major enteric pathogen responsible for antibiotic-associated diarrhea. Host susceptibility to C. difficile infections results partly from inability of the intestinal microbiota to resist C. difficile colonization. During early infancy, asymptomatic colonization by C. difficile is common and the intestinal microbiota shows low complexity. Thus, we investigated the potential relationship between the microbiota composition and the implantation of C. difficile in infant gut. Fecal samples from 53 infants, ages 0 to 13 months, 27 negative and 26 positive for C. difficile, were studied. Dominant microbiota profiles were assessed by PCR-temporal temperature gradient gel electrophoresis (TTGE). Bacterial signatures of the intestinal microbiota associated with colonization by C. difficile were deciphered using principal component analysis (PCA). Resulting bands of interest in TTGE profiles were excised, sequenced, and analyzed by nucleotide BLAST (NCBI). While global biodiversity was not affected, interclass PCA on instrumental variables highlighted significant differences in dominant bacterial species between C. difficile-colonized and noncolonized infants (P = 0.017). Four bands were specifically associated with the presence or absence of C. difficile: 16S rRNA gene sequences related to Ruminococcus gnavus and Klebsiella pneumoniae for colonized infants and to Bifidobacterium longum for noncolonized infants. We demonstrated that the presence of C. difficile in the intestinal microbiota of infants was associated with changes in this ecosystem's composition. These results suggest that the composition of the gut microbiota might be crucial in the colonization process, although the chronology of events remains to be determined.
Figures



References
-
- Bartlett J. G., Chang T. W., Gurwith M., Gorbach S. L., Onderdonk A. B. 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N. Engl. J. Med. 298:531–534 - PubMed
-
- Bartlett J. G. 2002. Clostridium difficile-associated enteric disease. Curr. Infect. Dis. Rep. 4:477–483 - PubMed
-
- Chang J. Y., et al. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197:435–438 - PubMed
-
- Collignon A., et al. 1993. Heterogeneity of Clostridium difficile isolates from infants. Eur. J. Pediatr. 152:319–322 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

