Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008
- PMID: 21177926
- PMCID: PMC3133703
- DOI: 10.1099/vir.0.027557-0
Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008
Abstract
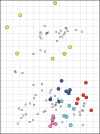
Prior to the introduction of the 2009 pandemic H1N1 virus from humans into pigs, four phylogenetic clusters (α-, β-, γ- and δ) of the haemagglutinin (HA) gene from H1 influenza viruses could be found in US swine. Information regarding the antigenic relatedness of the H1 viruses was lacking due to the dynamic and variable nature of swine lineage H1. We characterized 12 H1 isolates from 2008 by using 454 genome-sequencing technology and phylogenetic analysis of all eight gene segments and by serological cross-reactivity in the haemagglutination inhibition (HI) assay. Genetic diversity was demonstrated in all gene segments, but most notably in the HA gene. The gene segments from the 2009 pandemic H1N1 formed clusters separate from North American swine lineage viruses, suggesting progenitors of the pandemic virus were not present in US pigs immediately prior to 2009. Serological cross-reactivity paired with antigenic cartography demonstrated that the viruses in the different phylogenetic clusters are also antigenically divergent.
Figures



References
-
- Chen, W., Calvo, P. A., Malide, D., Gibbs, J., Schubert, U., Bacik, I., Basta, S., O'Neill, R., Schickli, J. & other authors (2001). A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7, 1306–1312. - PubMed
-
- Dawood, F. S., Jain, S., Finelli, L., Shaw, M. W., Lindstrom, S., Garten, R. J., Gubareva, L. V., Xu, X., Bridges, C. B. & other authors (2009). Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 360, 2605–2615. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

