A global analysis of C. elegans trans-splicing
- PMID: 21177958
- PMCID: PMC3032929
- DOI: 10.1101/gr.113811.110
A global analysis of C. elegans trans-splicing
Abstract
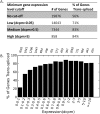
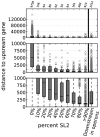
Trans-splicing of one of two short leader RNAs, SL1 or SL2, occurs at the 5' ends of pre-mRNAs of many C. elegans genes. We have exploited RNA-sequencing data from the modENCODE project to analyze the transcriptome of C. elegans for patterns of trans-splicing. Transcripts of ∼70% of genes are trans-spliced, similar to earlier estimates based on analysis of far fewer genes. The mRNAs of most trans-spliced genes are spliced to either SL1 or SL2, but most genes are not trans-spliced to both, indicating that SL1 and SL2 trans-splicing use different underlying mechanisms. SL2 trans-splicing occurs in order to separate the products of genes in operons genome wide. Shorter intercistronic distance is associated with greater use of SL2. Finally, increased use of SL1 trans-splicing to downstream operon genes can indicate the presence of an extra promoter in the intercistronic region, creating what has been termed a "hybrid" operon. Within hybrid operons the presence of the two promoters results in the use of the two SL classes: Transcription that originates at the promoter upstream of another gene creates a polycistronic pre-mRNA that receives SL2, whereas transcription that originates at the internal promoter creates transcripts that receive SL1. Overall, our data demonstrate that >17% of all C. elegans genes are in operons.
Figures

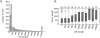
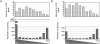

References
-
- Beißbarth T, Speed TP 2004. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20: 1464–1465 - PubMed
-
- Blumenthal T 2005. Trans-splicing and operons. WormBook 25: 1–9 - PubMed
-
- Blumenthal T, Spieth J 1996. Gene structure and organization in Caenorhabditis elegans. Curr Opin Genet Dev 6: 692–698 - PubMed
-
- Blumenthal T, Steward K 1997. Trans-splicing. In C. elegans II, pp. 129–132 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
