Genome-wide analysis of promoter architecture in Drosophila melanogaster
- PMID: 21177961
- PMCID: PMC3032922
- DOI: 10.1101/gr.112466.110
Genome-wide analysis of promoter architecture in Drosophila melanogaster
Abstract
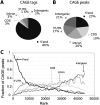
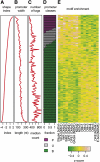
Core promoters are critical regions for gene regulation in higher eukaryotes. However, the boundaries of promoter regions, the relative rates of initiation at the transcription start sites (TSSs) distributed within them, and the functional significance of promoter architecture remain poorly understood. We produced a high-resolution map of promoters active in the Drosophila melanogaster embryo by integrating data from three independent and complementary methods: 21 million cap analysis of gene expression (CAGE) tags, 1.2 million RNA ligase mediated rapid amplification of cDNA ends (RLM-RACE) reads, and 50,000 cap-trapped expressed sequence tags (ESTs). We defined 12,454 promoters of 8037 genes. Our analysis indicates that, due to non-promoter-associated RNA background signal, previous studies have likely overestimated the number of promoter-associated CAGE clusters by fivefold. We show that TSS distributions form a complex continuum of shapes, and that promoters active in the embryo and adult have highly similar shapes in 95% of cases. This suggests that these distributions are generally determined by static elements such as local DNA sequence and are not modulated by dynamic signals such as histone modifications. Transcription factor binding motifs are differentially enriched as a function of promoter shape, and peaked promoter shape is correlated with both temporal and spatial regulation of gene expression. Our results contribute to the emerging view that core promoters are functionally diverse and control patterning of gene expression in Drosophila and mammals.
Figures




References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 - PubMed
-
- Bickel PJ, Brown JB, Boley N, Huang H, Zhang N 2011. Non parametric methods for genomic inference. Ann Appl Stat (in press)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
