Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin
- PMID: 21177972
- PMCID: PMC3032919
- DOI: 10.1101/gr.110098.110
Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin
Abstract
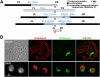
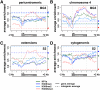
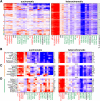
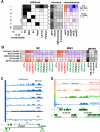
Eukaryotic genomes are packaged in two basic forms, euchromatin and heterochromatin. We have examined the composition and organization of Drosophila melanogaster heterochromatin in different cell types using ChIP-array analysis of histone modifications and chromosomal proteins. As anticipated, the pericentric heterochromatin and chromosome 4 are on average enriched for the "silencing" marks H3K9me2, H3K9me3, HP1a, and SU(VAR)3-9, and are generally depleted for marks associated with active transcription. The locations of the euchromatin-heterochromatin borders identified by these marks are similar in animal tissues and most cell lines, although the amount of heterochromatin is variable in some cell lines. Combinatorial analysis of chromatin patterns reveals distinct profiles for euchromatin, pericentric heterochromatin, and the 4th chromosome. Both silent and active protein-coding genes in heterochromatin display complex patterns of chromosomal proteins and histone modifications; a majority of the active genes exhibit both "activation" marks (e.g., H3K4me3 and H3K36me3) and "silencing" marks (e.g., H3K9me2 and HP1a). The hallmark of active genes in heterochromatic domains appears to be a loss of H3K9 methylation at the transcription start site. We also observe complex epigenomic profiles of intergenic regions, repeated transposable element (TE) sequences, and genes in the heterochromatic extensions. An unexpectedly large fraction of sequences in the euchromatic chromosome arms exhibits a heterochromatic chromatin signature, which differs in size, position, and impact on gene expression among cell types. We conclude that patterns of heterochromatin/euchromatin packaging show greater complexity and plasticity than anticipated. This comprehensive analysis provides a foundation for future studies of gene activity and chromosomal functions that are influenced by or dependent upon heterochromatin.
Figures





References
-
- Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ 3rd 2003. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes Dev 17: 1855–1869 - PMC - PubMed
-
- Belyaeva ES, Andreyeva EN, Belyakin SN, Volkova EI, Zhimulev IF 2008. Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster. Chromosoma 117: 411–418 - PubMed
-
- Belyakin SN, Christophides GK, Alekseyenko AA, Kriventseva EV, Belyaeva ES, Nanayev RA, Makunin IV, Kafatos FC, Zhimulev IF 2005. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc Natl Acad Sci 102: 8269–8274 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
