A novel role for IFN-stimulated gene factor 3II in IFN-γ signaling and induction of antiviral activity in human cells
- PMID: 21178011
- PMCID: PMC3417313
- DOI: 10.4049/jimmunol.1001359
A novel role for IFN-stimulated gene factor 3II in IFN-γ signaling and induction of antiviral activity in human cells
Abstract
Type I (e.g., IFN-α, IFN-β) and type II IFNs (IFN-γ) have antiviral, antiproliferative, and immunomodulatory properties. Both types of IFN signal through the Jak/STAT pathway to elicit antiviral activity, yet IFN-γ is thought to do so only through STAT1 homodimers, whereas type I IFNs activate both STAT1- and STAT2-containing complexes such as IFN-stimulated gene factor 3. In this study, we show that IFN-stimulated gene factor 3 containing unphosphorylated STAT2 (ISGF3(II)) also plays a role in IFN-γ-mediated antiviral activity in humans. Using phosphorylated STAT1 as a marker for IFN signaling, Western blot analysis of IFN-α2a-treated human A549 cells revealed that phospho-STAT1 (Y701) levels peaked at 1 h, decreased by 6 h, and remained at low levels for up to 48 h. Cells treated with IFN-γ showed a biphasic phospho-STAT1 response with an early peak at 1-2 h and a second peak at 15-24 h. Gene expression microarray following IFN-γ treatment for 24 h indicated an induction of antiviral genes that are induced by IFN-stimulated gene factor 3 and associated with a type I IFN response. Induction of these genes by autocrine type I and type III IFN signaling was ruled out using neutralizing Abs to these IFNs in biological assays and by quantitative RT-PCR. Despite the absence of autocrine IFNs, IFN-γ treatment induced formation of ISGF3(II). This novel transcription factor complex binds to IFN-stimulated response element promoter sequences, as shown by chromatin immunoprecipitation analysis of the protein kinase R promoter. STAT2 and IFN regulatory factor 9 knockdown in A549 cells reversed IFN-γ-mediated IFN-stimulated response element induction and antiviral activity, implicating ISGF3(II) formation as a significant component of the cellular response and biological activity of IFN-γ.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures






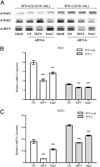

References
-
- Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annu. Rev. Biochem. 1987;56:727–777. - PubMed
-
- Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell. Microbiol. 2006;8:907–922. - PubMed
-
- Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. Interferon: cellular executioner or white knight? Curr. Med. Chem. 2007;14:1279–1289. - PubMed
-
- Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 1997;15:563–591. - PubMed
-
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

