Metabonomic, transcriptomic, and genomic variation of a population cohort
- PMID: 21179014
- PMCID: PMC3018170
- DOI: 10.1038/msb.2010.93
Metabonomic, transcriptomic, and genomic variation of a population cohort
Abstract
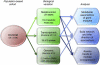
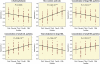
Comprehensive characterization of human tissues promises novel insights into the biological architecture of human diseases and traits. We assessed metabonomic, transcriptomic, and genomic variation for a large population-based cohort from the capital region of Finland. Network analyses identified a set of highly correlated genes, the lipid-leukocyte (LL) module, as having a prominent role in over 80 serum metabolites (of 134 measures quantified), including lipoprotein subclasses, lipids, and amino acids. Concurrent association with immune response markers suggested the LL module as a possible link between inflammation, metabolism, and adiposity. Further, genomic variation was used to generate a directed network and infer LL module's largely reactive nature to metabolites. Finally, gene co-expression in circulating leukocytes was shown to be dependent on serum metabolite concentrations, providing evidence for the hypothesis that the coherence of molecular networks themselves is conditional on environmental factors. These findings show the importance and opportunity of systematic molecular investigation of human population samples. To facilitate and encourage this investigation, the metabonomic, transcriptomic, and genomic data used in this study have been made available as a resource for the research community.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Ala-Korpela M (1995) 1H NMR spectroscopy of human blood plasma. Prog Nucl Magn Reson Spectr 27: 475–554
-
- Ala-Korpela M (2008) Critical evaluation of 1H NMR metabonomics of serum as a methodology for disease risk assessment and diagnostics. Clin Chem Lab Med 46: 27–42 - PubMed
-
- Ala-Korpela M, Soininen P, Savolainen MJ (2009) Letter by Ala-Korpela et al regarding article, ‘Lipoprotein particle profiles by nuclear magnetic resonance compared with standard lipids and apolipoproteins in predicting incident cardiovascular disease in women’. Circulation 120: e149; author reply e150 - PubMed
-
- Altmann F (2007) The role of protein glycosylation in allergy. Int Arch Allergy Immunol 142: 99–115 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

