Defining the budding yeast chromatin-associated interactome
- PMID: 21179020
- PMCID: PMC3018163
- DOI: 10.1038/msb.2010.104
Defining the budding yeast chromatin-associated interactome
Abstract
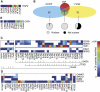
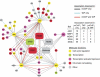
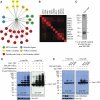
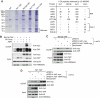
We previously reported a novel affinity purification (AP) method termed modified chromatin immunopurification (mChIP), which permits selective enrichment of DNA-bound proteins along with their associated protein network. In this study, we report a large-scale study of the protein network of 102 chromatin-related proteins from budding yeast that were analyzed by mChIP coupled to mass spectrometry. This effort resulted in the detection of 2966 high confidence protein associations with 724 distinct preys. mChIP resulted in significantly improved interaction coverage as compared with classical AP methodology for ∼75% of the baits tested. Furthermore, mChIP successfully identified novel binding partners for many lower abundance transcription factors that previously failed using conventional AP methodologies. mChIP was also used to perform targeted studies, particularly of Asf1 and its associated proteins, to allow for a understanding of the physical interplay between Asf1 and two other histone chaperones, Rtt106 and the HIR complex, to be gained.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

