Defining the transcriptome and proteome in three functionally different human cell lines
- PMID: 21179022
- PMCID: PMC3018165
- DOI: 10.1038/msb.2010.106
Defining the transcriptome and proteome in three functionally different human cell lines
Abstract
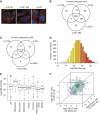
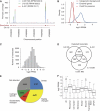
An essential question in human biology is how cells and tissues differ in gene and protein expression and how these differences delineate specific biological function. Here, we have performed a global analysis of both mRNA and protein levels based on sequence-based transcriptome analysis (RNA-seq), SILAC-based mass spectrometry analysis and antibody-based confocal microscopy. The study was performed in three functionally different human cell lines and based on the global analysis, we estimated the fractions of mRNA and protein that are cell specific or expressed at similar/different levels in the cell lines. A highly ubiquitous RNA expression was found with >60% of the gene products detected in all cells. The changes of mRNA and protein levels in the cell lines using SILAC and RNA ratios show high correlations, even though the genome-wide dynamic range is substantially higher for the proteins as compared with the transcripts. Large general differences in abundance for proteins from various functional classes are observed and, in general, the cell-type specific proteins are low abundant and highly enriched for cell-surface proteins. Thus, this study shows a path to characterize the transcriptome and proteome in human cells from different origins.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
-
- Barbe L, Lundberg E, Oksvold P, Stenius A, Lewin E, Bjorling E, Asplund A, Ponten F, Brismar H, Uhlen M, Andersson-Svahn H (2008) Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics 7: 499–508 - PubMed
-
- Berglund L, Bjorling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernerus H, Nilsson P, Lundberg E, Sivertsson A, Navani S, Wester K, Kampf C, Hober S, Ponten F, Uhlen M (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics 7: 2019–2027 - PubMed
-
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 - PMC - PubMed
-
- Bjorling E, Uhlen M (2008) Antibodypedia, a portal for sharing antibody and antigen validation data. Mol Cell Proteomics 7: 2028–2037 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

