Mechanisms of mitophagy
- PMID: 21179058
- PMCID: PMC4780047
- DOI: 10.1038/nrm3028
Mechanisms of mitophagy
Abstract
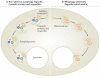
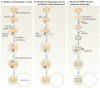
Autophagy not only recycles intracellular components to compensate for nutrient deprivation but also selectively eliminates organelles to regulate their number and maintain quality control. Mitophagy, the specific autophagic elimination of mitochondria, has been identified in yeast, mediated by autophagy-related 32 (Atg32), and in mammals during red blood cell differentiation, mediated by NIP3-like protein X (NIX; also known as BNIP3L). Moreover, mitophagy is regulated in many metazoan cell types by parkin and PTEN-induced putative kinase protein 1 (PINK1), and mutations in the genes encoding these proteins have been linked to forms of Parkinson's disease.
Figures
References
-
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature Rev. Mol. Cell Biol. 2009;10:458–467. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

