An autoimmune-associated variant in PTPN2 reveals an impairment of IL-2R signaling in CD4(+) T cells
- PMID: 21179116
- PMCID: PMC3058680
- DOI: 10.1038/gene.2010.54
An autoimmune-associated variant in PTPN2 reveals an impairment of IL-2R signaling in CD4(+) T cells
Abstract
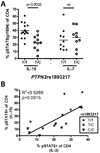
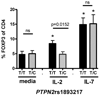
The IL-2/IL-2R signaling pathway has an important role in autoimmunity. Several genes identified in genome-wide association (GWA) studies encode proteins in the IL-2/IL-2R signaling cascade that are associated with autoimmune diseases. One of these, PTPN2, encodes a protein tyrosine phosphatase that is highly expressed in T cells and regulates cytokine signaling. An intronic risk allele in PTPN2, rs1893217(C), correlated with decreased IL-2R signaling in CD4(+) T cells as measured by phosphorylation of STAT5 (phosphorylated STAT5 (pSTAT5)). We modeled an additive single nucleotide polymorphism (SNP) genotype, in which each copy of the risk allele conferred a decrease in IL-2R signaling (P=4.4 × 10(-8)). Decreased pSTAT5 impacted IL-2Rβ chain signaling resulting in reduced FOXP3 expression in activated cells. This phenotype was not due to overt differences in expression of the IL-2R, molecules in the IL-2R signaling cascade or defects in STAT5. However, the rs1893217(C) risk variant did correlate with decreased PTPN2 expression in CD4(+)CD45RO T cells (P=0.0002). Thus, the PTPN2rs1893217(C) risk allele associated with reduced pSTAT5 in response to IL-2 and reduced PTPN2 expression. Together, these data suggest that decreased expression of PTPN2 may indirectly modulate IL-2 responsiveness. These findings, identified through genotype/phenotype relationships, may lead to identification of novel mechanisms underlying dysregulation of cytokine signaling in autoimmunity.
Conflict of interest statement
Figures



References
-
- Sadlack B, Lohler J, Schorle H, Klebb G, Haber H, Sickel E, et al. Generalized autoimmune disease in interleukin-2-deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur J Immunol. 1995 Nov;25(11):3053–3059. - PubMed
-
- Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, et al. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 1995 Jun 9;268(5216):1472–1476. - PubMed
-
- Hakonarson H, Qu HQ, Bradfield JP, Marchand L, Kim CE, Glessner JT, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008 Apr;57(4):1143–1146. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

