Accurate protein structure annotation through competitive diffusion of enzymatic functions over a network of local evolutionary similarities
- PMID: 21179190
- PMCID: PMC3001439
- DOI: 10.1371/journal.pone.0014286
Accurate protein structure annotation through competitive diffusion of enzymatic functions over a network of local evolutionary similarities
Abstract
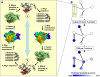
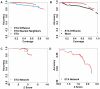
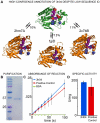
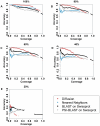
High-throughput Structural Genomics yields many new protein structures without known molecular function. This study aims to uncover these missing annotations by globally comparing select functional residues across the structural proteome. First, Evolutionary Trace Annotation, or ETA, identifies which proteins have local evolutionary and structural features in common; next, these proteins are linked together into a proteomic network of ETA similarities; then, starting from proteins with known functions, competing functional labels diffuse link-by-link over the entire network. Every node is thus assigned a likelihood z-score for every function, and the most significant one at each node wins and defines its annotation. In high-throughput controls, this competitive diffusion process recovered enzyme activity annotations with 99% and 97% accuracy at half-coverage for the third and fourth Enzyme Commission (EC) levels, respectively. This corresponds to false positive rates 4-fold lower than nearest-neighbor and 5-fold lower than sequence-based annotations. In practice, experimental validation of the predicted carboxylesterase activity in a protein from Staphylococcus aureus illustrated the effectiveness of this approach in the context of an increasingly drug-resistant microbe. This study further links molecular function to a small number of evolutionarily important residues recognizable by Evolutionary Tracing and it points to the specificity and sensitivity of functional annotation by competitive global network diffusion. A web server is at http://mammoth.bcm.tmc.edu/networks.
Conflict of interest statement
Figures


References
-
- Friedberg I. Automated protein function prediction--the genomic challenge. Brief Bioinform. 2006;7:225–242. - PubMed
-
- Chandonia J, Brenner SE. The Impact of Structural Genomics: Expectations and Outcomes. Science. 2006;311:347–351. - PubMed
-
- Brenner SE. Errors in genome annotation. Trends in Genetics. 1999;15:132–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

