The PluriNetWork: an electronic representation of the network underlying pluripotency in mouse, and its applications
- PMID: 21179244
- PMCID: PMC3003487
- DOI: 10.1371/journal.pone.0015165
The PluriNetWork: an electronic representation of the network underlying pluripotency in mouse, and its applications
Abstract
Background: Analysis of the mechanisms underlying pluripotency and reprogramming would benefit substantially from easy access to an electronic network of genes, proteins and mechanisms. Moreover, interpreting gene expression data needs to move beyond just the identification of the up-/downregulation of key genes and of overrepresented processes and pathways, towards clarifying the essential effects of the experiment in molecular terms.
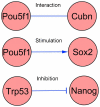
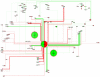
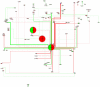
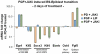
Methodology/principal findings: We have assembled a network of 574 molecular interactions, stimulations and inhibitions, based on a collection of research data from 177 publications until June 2010, involving 274 mouse genes/proteins, all in a standard electronic format, enabling analyses by readily available software such as Cytoscape and its plugins. The network includes the core circuit of Oct4 (Pou5f1), Sox2 and Nanog, its periphery (such as Stat3, Klf4, Esrrb, and c-Myc), connections to upstream signaling pathways (such as Activin, WNT, FGF, BMP, Insulin, Notch and LIF), and epigenetic regulators as well as some other relevant genes/proteins, such as proteins involved in nuclear import/export. We describe the general properties of the network, as well as a Gene Ontology analysis of the genes included. We use several expression data sets to condense the network to a set of network links that are affected in the course of an experiment, yielding hypotheses about the underlying mechanisms.
Conclusions/significance: We have initiated an electronic data repository that will be useful to understand pluripotency and to facilitate the interpretation of high-throughput data. To keep up with the growth of knowledge on the fundamental processes of pluripotency and reprogramming, we suggest to combine Wiki and social networking software towards a community curation system that is easy to use and flexible, and tailored to provide a benefit for the scientist, and to improve communication and exchange of research results. A PluriNetWork tutorial is available at http://www.ibima.med.uni-rostock.de/IBIMA/PluriNetWork/.
Conflict of interest statement
Figures




References
-
- Do JT, Scholer HR. Regulatory circuits underlying pluripotency and reprogramming. Trends Pharmacol Sci. 2009;30:296–302. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

