A dual platform approach to transcript discovery for the planarian Schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology
- PMID: 21179477
- PMCID: PMC3001875
- DOI: 10.1371/journal.pone.0015617
A dual platform approach to transcript discovery for the planarian Schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology
Abstract
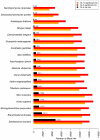
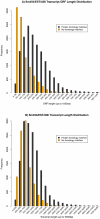
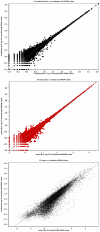
The use of planarians as a model system is expanding and the mechanisms that control planarian regeneration are being elucidated. The planarian Schmidtea mediterranea in particular has become a species of choice. Currently the planarian research community has access to this whole genome sequencing project and over 70,000 expressed sequence tags. However, the establishment of massively parallel sequencing technologies has provided the opportunity to define genetic content, and in particular transcriptomes, in unprecedented detail. Here we apply this approach to the planarian model system. We have sequenced, mapped and assembled 581,365 long and 507,719,814 short reads from RNA of intact and mixed stages of the first 7 days of planarian regeneration. We used an iterative mapping approach to identify and define de novo splice sites with short reads and increase confidence in our transcript predictions. We more than double the number of transcripts currently defined by publicly available ESTs, resulting in a collection of 25,053 transcripts described by combining platforms. We also demonstrate the utility of this collection for an RNAseq approach to identify potential transcripts that are enriched in neoblast stem cells and their progeny by comparing transcriptome wide expression levels between irradiated and intact planarians. Our experiments have defined an extensive planarian transcriptome that can be used as a template for RNAseq and can also help to annotate the S. mediterranea genome. We anticipate that suites of other 'omic approaches will also be facilitated by building on this comprehensive data set including RNAseq across many planarian regenerative stages, scenarios, tissues and phenotypes generated by RNAi.
Conflict of interest statement
Figures







References
-
- Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–549. - PubMed
-
- Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. - PubMed
-
- Salo E. The power of regeneration and the stem-cell kingdom: freshwater planarians (Platyhelminthes). Bioessays. 2006;28:546–559. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

