Dispersal of biofilms by secreted, matrix degrading, bacterial DNase
- PMID: 21179489
- PMCID: PMC3001887
- DOI: 10.1371/journal.pone.0015668
Dispersal of biofilms by secreted, matrix degrading, bacterial DNase
Abstract
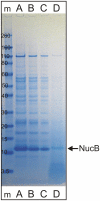
Microbial biofilms are composed of a hydrated matrix of biopolymers including polypeptides, polysaccharides and nucleic acids and act as a protective barrier and microenvironment for the inhabiting microbes. While studying marine biofilms, we observed that supernatant produced by a marine isolate of Bacillus licheniformis was capable of dispersing bacterial biofilms. We investigated the source of this activity and identified the active compound as an extracellular DNase (NucB). We have shown that this enzyme rapidly breaks up the biofilms of both Gram-positive and Gram-negative bacteria. We demonstrate that bacteria can use secreted nucleases as an elegant strategy to disperse established biofilms and to prevent de novo formation of biofilms of competitors. DNA therefore plays an important dynamic role as a reversible structural adhesin within the biofilm.
Conflict of interest statement
Figures




References
-
- Fletcher M. Bacterial biofilms and biofouling. Curr Opin Biotechnol. 1994;5:302–306. - PubMed
-
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. - PubMed
-
- Costerton JW. Overview of microbial biofilms. J Ind Microbiol. 1995;15:137–140. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

