Polyploidization altered gene functions in cotton (Gossypium spp.)
- PMID: 21179551
- PMCID: PMC3002935
- DOI: 10.1371/journal.pone.0014351
Polyploidization altered gene functions in cotton (Gossypium spp.)
Abstract
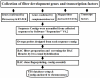
Cotton (Gossypium spp.) is an important crop plant that is widely grown to produce both natural textile fibers and cottonseed oil. Cotton fibers, the economically more important product of the cotton plant, are seed trichomes derived from individual cells of the epidermal layer of the seed coat. It has been known for a long time that large numbers of genes determine the development of cotton fiber, and more recently it has been determined that these genes are distributed across At and Dt subgenomes of tetraploid AD cottons. In the present study, the organization and evolution of the fiber development genes were investigated through the construction of an integrated genetic and physical map of fiber development genes whose functions have been verified and confirmed. A total of 535 cotton fiber development genes, including 103 fiber transcription factors, 259 fiber development genes, and 173 SSR-contained fiber ESTs, were analyzed at the subgenome level. A total of 499 fiber related contigs were selected and assembled. Together these contigs covered about 151 Mb in physical length, or about 6.7% of the tetraploid cotton genome. Among the 499 contigs, 397 were anchored onto individual chromosomes. Results from our studies on the distribution patterns of the fiber development genes and transcription factors between the At and Dt subgenomes showed that more transcription factors were from Dt subgenome than At, whereas more fiber development genes were from At subgenome than Dt. Combining our mapping results with previous reports that more fiber QTLs were mapped in Dt subgenome than At subgenome, the results suggested a new functional hypothesis for tetraploid cotton. After the merging of the two diploid Gossypium genomes, the At subgenome has provided most of the genes for fiber development, because it continues to function similar to its fiber producing diploid A genome ancestor. On the other hand, the Dt subgenome, with its non-fiber producing D genome ancestor, provides more transcription factors that regulate the expression of the fiber genes in the At subgenome. This hypothesis would explain previously published mapping results. At the same time, this integrated map of fiber development genes would provide a framework to clone individual full-length fiber genes, to elucidate the physiological mechanisms of the fiber differentiation, elongation, and maturation, and to systematically study the functional network of these genes that interact during the process of fiber development in the tetraploid cottons.
Conflict of interest statement
Figures


References
-
- Fryxell PA. A revised taxonomic interpretation of Gossypium L.(Malvaceae). Rheeda. 1992;2:108–165.
-
- Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186.
-
- Ulloa M, Saha S, Jenkins JN, Meredith WR, Jr, McCarty JC, Jr, et al. Chromosomal assignment of RFLP linkage groups harboring important QTLs on an intraspecific cotton (Gossypium hirsutum L.) Joinmap. J Hered. 2005;96:132–144. - PubMed
-
- Applequist WL, Cronn R, Wendel JF. Comparative development of fiber in wild and cultivated cotton. Evol Dev. 2001;3:3–17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

