Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins
- PMID: 21183075
- PMCID: PMC3472970
- DOI: 10.1016/j.cell.2010.11.042
Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins
Abstract
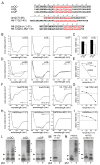
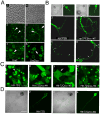
The functional switch of glutamine/asparagine (Q/N)-rich prions and the neurotoxicity of polyQ-expanded proteins involve complex aggregation-prone structural transitions, commonly presumed to be forming β sheets. By analyzing sequences of interaction partners of these proteins, we discovered a recurrent presence of coiled-coil domains both in the partners and in segments that flank or overlap Q/N-rich and polyQ domains. Since coiled coils can mediate protein interactions and multimerization, we studied their possible involvement in Q/N-rich and polyQ aggregations. Using circular dichroism and chemical crosslinking, we found that Q/N-rich and polyQ peptides form α-helical coiled coils in vitro and assemble into multimers. Using structure-guided mutagenesis, we found that coiled-coil domains modulate in vivo properties of two Q/N-rich prions and polyQ-expanded huntingtin. Mutations that disrupt coiled coils impair aggregation and activity, whereas mutations that enhance coiled-coil propensity promote aggregation. These findings support a coiled-coil model for the functional switch of Q/N-rich prions and for the pathogenesis of polyQ-expansion diseases.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures





References
-
- Bussell R, Jr, Eliezer D. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J Mol Biol. 2003;329:763–778. - PubMed
-
- Chang DK, Cheng SF, Trivedi VD, Lin KL. Proline affects oligomerization of a coiled coil by inducing a kink in a long helix. J Struct Biol. 1999;128:270–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

