Metagenomic analysis of the viral communities in fermented foods
- PMID: 21183634
- PMCID: PMC3067239
- DOI: 10.1128/AEM.01859-10
Metagenomic analysis of the viral communities in fermented foods
Abstract
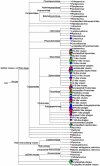
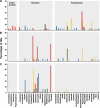
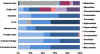
Viruses are recognized as the most abundant biological components on Earth, and they regulate the structure of microbial communities in many environments. In soil and marine environments, microorganism-infecting phages are the most common type of virus. Although several types of bacteriophage have been isolated from fermented foods, little is known about the overall viral assemblages (viromes) of these environments. In this study, metagenomic analyses were performed on the uncultivated viral communities from three fermented foods, fermented shrimp, kimchi, and sauerkraut. Using a high-throughput pyrosequencing technique, a total of 81,831, 70,591 and 69,464 viral sequences were obtained from fermented shrimp, kimchi and sauerkraut, respectively. Moreover, 37 to 50% of these sequences showed no significant hit against sequences in public databases. There were some discrepancies between the prediction of bacteriophages hosts via homology comparison and bacterial distribution, as determined from 16S rRNA gene sequencing. These discrepancies likely reflect the fact that the viral genomes of fermented foods are poorly represented in public databases. Double-stranded DNA viral communities were amplified from fermented foods by using a linker-amplified shotgun library. These communities were dominated by bacteriophages belonging to the viral order Caudovirales (i.e., Myoviridae, Podoviridae, and Siphoviridae). This study indicates that fermented foods contain less complex viral communities than many other environmental habitats, such as seawater, human feces, marine sediment, and soil.
Figures
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

