Optimizing non-natural protein function with directed evolution
- PMID: 21185770
- PMCID: PMC3080047
- DOI: 10.1016/j.cbpa.2010.11.020
Optimizing non-natural protein function with directed evolution
Abstract
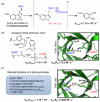
Developing technologies such as unnatural amino acid mutagenesis, non-natural cofactor engineering, and computational design are generating proteins with novel functions; these proteins, however, often do not reach performance targets and would benefit from further optimization. Evolutionary methods can complement these approaches: recent work combining unnatural amino acid mutagenesis and phage selection has created useful proteins of novel composition. Weak initial activity in a computationally designed enzyme has been improved by iterative rounds of mutagenesis and screening. A marriage of ingenuity and evolution will expand the scope of protein function well beyond Mother Nature's designs.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures

References
-
- Turner NJ. Directed evolution drives the next generation of biocatalysts. Nat Chem Biol. 2009;5:567–573. - PubMed
-
- Jackel C, Hilvert D. Biocatalysts by evolution. Curr Opin Biotechnol. 2010;21 - PubMed
-
- Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. - PubMed
-
- Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

