A mammalian functional-genetic approach to characterizing cancer therapeutics
- PMID: 21186347
- PMCID: PMC3070540
- DOI: 10.1038/nchembio.503
A mammalian functional-genetic approach to characterizing cancer therapeutics
Abstract
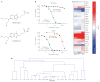
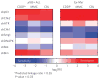
Identifying mechanisms of drug action remains a fundamental impediment to the development and effective use of chemotherapeutics. Here we describe an RNA interference (RNAi)-based strategy to characterize small-molecule function in mammalian cells. By examining the response of cells expressing short hairpin RNAs (shRNAs) to a diverse selection of chemotherapeutics, we could generate a functional shRNA signature that was able to accurately group drugs into established biochemical modes of action. This, in turn, provided a diversely sampled reference set for high-resolution prediction of mechanisms of action for poorly characterized small molecules. We could further reduce the predictive shRNA target set to as few as eight genes and, by using a newly derived probability-based nearest-neighbors approach, could extend the predictive power of this shRNA set to characterize additional drug categories. Thus, a focused shRNA phenotypic signature can provide a highly sensitive and tractable approach for characterizing new anticancer drugs.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Sato S, Murata A, Shirakawa T, Uesugi M. Biochemical target isolation for novices: affinity-based strategies. Chem Biol. 2010;17:616–623. - PubMed
-
- Giaever G, et al. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet. 1999;21:278–283. - PubMed
-
- Lum PY, et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. - PubMed
-
- Parsons AB, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

