The regulation of autophagy - unanswered questions
- PMID: 21187343
- PMCID: PMC3037094
- DOI: 10.1242/jcs.064576
The regulation of autophagy - unanswered questions
Abstract
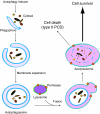
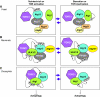
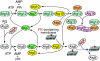
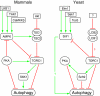
Autophagy is an intracellular lysosomal (vacuolar) degradation process that is characterized by the formation of double-membrane vesicles, known as autophagosomes, which sequester cytoplasm. As autophagy is involved in cell growth, survival, development and death, the levels of autophagy must be properly regulated, as indicated by the fact that dysregulated autophagy has been linked to many human pathophysiologies, such as cancer, myopathies, neurodegeneration, heart and liver diseases, and gastrointestinal disorders. Substantial progress has recently been made in understanding the molecular mechanisms of the autophagy machinery, and in the regulation of autophagy. However, many unanswered questions remain, such as how the Atg1 complex is activated and the function of PtdIns3K is regulated, how the ubiquitin-like conjugation systems participate in autophagy and the mechanisms of phagophore expansion and autophagosome formation, how the network of TOR signaling pathways regulating autophagy are controlled, and what the underlying mechanisms are for the pro-cell survival and the pro-cell death effects of autophagy. As several recent reviews have comprehensively summarized the recent progress in the regulation of autophagy, we focus in this Commentary on the main unresolved questions in this field.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

