Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast
- PMID: 21187411
- PMCID: PMC3021021
- DOI: 10.1073/pnas.1017570108
Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast
Abstract
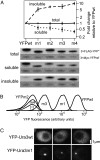
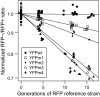
Evolving lineages face a constant intracellular threat: most new coding sequence mutations destabilize the folding of the encoded protein. Misfolded proteins form insoluble aggregates and are hypothesized to be intrinsically cytotoxic. Here, we experimentally isolate a fitness cost caused by toxicity of misfolded proteins. We exclude other costs of protein misfolding, such as loss of functional protein or attenuation of growth-limiting protein synthesis resources, by comparing growth rates of budding yeast expressing folded or misfolded variants of a gratuitous protein, YFP, at equal levels. We quantify a fitness cost that increases with misfolded protein abundance, up to as much as a 3.2% growth rate reduction when misfolded YFP represents less than 0.1% of total cellular protein. Comparable experiments on variants of the yeast gene orotidine-5'-phosphate decarboxylase (URA3) produce similar results. Quantitative proteomic measurements reveal that, within the cell, misfolded YFP induces coordinated synthesis of interacting cytosolic chaperone proteins in the absence of a wider stress response, providing evidence for an evolved modular response to misfolded proteins in the cytosol. These results underscore the distinct and evolutionarily relevant molecular threat of protein misfolding, independent of protein function. Assuming that most misfolded proteins impose similar costs, yeast cells express almost all proteins at steady-state levels sufficient to expose their encoding genes to selection against misfolding, lending credibility to the recent suggestion that such selection imposes a global constraint on molecular evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Pakula AA, Sauer RT. Genetic analysis of protein stability and function. Annu Rev Genet. 1989;23:289–310. - PubMed
-
- Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. - PubMed
-
- Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–699. - PubMed
-
- Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

