H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3
- PMID: 21187892
- PMCID: PMC3002978
- DOI: 10.1371/journal.ppat.1001223
H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3
Abstract
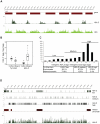
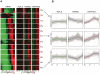
Epigenetic regulatory mechanisms and their enzymes are promising targets for malaria therapeutic intervention; however, the epigenetic component of gene expression in P. falciparum is poorly understood. Dynamic or stable association of epigenetic marks with genomic features provides important clues about their function and helps to understand how histone variants/modifications are used for indexing the Plasmodium epigenome. We describe a novel, linear amplification method for next-generation sequencing (NGS) that allows unbiased analysis of the extremely AT-rich Plasmodium genome. We used this method for high resolution, genome-wide analysis of a histone H2A variant, H2A.Z and two histone H3 marks throughout parasite intraerythrocytic development. Unlike in other organisms, H2A.Z is a constant, ubiquitous feature of euchromatic intergenic regions throughout the intraerythrocytic cycle. The almost perfect colocalisation of H2A.Z with H3K9ac and H3K4me3 suggests that these marks are preferentially deposited on H2A.Z-containing nucleosomes. By performing RNA-seq on 8 time-points, we show that acetylation of H3K9 at promoter regions correlates very well with the transcriptional status whereas H3K4me3 appears to have stage-specific regulation, being low at early stages, peaking at trophozoite stage, but does not closely follow changes in gene expression. Our improved NGS library preparation procedure provides a foundation to exploit the malaria epigenome in detail. Furthermore, our findings place H2A.Z at the cradle of P. falciparum epigenetic regulation by stably defining intergenic regions and providing a platform for dynamic assembly of epigenetic and other transcription related complexes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. - PubMed
-
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

