Comparative genomics suggests that the fungal pathogen pneumocystis is an obligate parasite scavenging amino acids from its host's lungs
- PMID: 21188143
- PMCID: PMC3004796
- DOI: 10.1371/journal.pone.0015152
Comparative genomics suggests that the fungal pathogen pneumocystis is an obligate parasite scavenging amino acids from its host's lungs
Abstract
Pneumocystis jirovecii is a fungus causing severe pneumonia in immuno-compromised patients. Progress in understanding its pathogenicity and epidemiology has been hampered by the lack of a long-term in vitro culture method. Obligate parasitism of this pathogen has been suggested on the basis of various features but remains controversial. We analysed the 7.0 Mb draft genome sequence of the closely related species Pneumocystis carinii infecting rats, which is a well established experimental model of the disease. We predicted 8'085 (redundant) peptides and 14.9% of them were mapped onto the KEGG biochemical pathways. The proteome of the closely related yeast Schizosaccharomyces pombe was used as a control for the annotation procedure (4'974 genes, 14.1% mapped). About two thirds of the mapped peptides of each organism (65.7% and 73.2%, respectively) corresponded to crucial enzymes for the basal metabolism and standard cellular processes. However, the proportion of P. carinii genes relative to those of S. pombe was significantly smaller for the "amino acid metabolism" category of pathways than for all other categories taken together (40 versus 114 against 278 versus 427, P<0.002). Importantly, we identified in P. carinii only 2 enzymes specifically dedicated to the synthesis of the 20 standard amino acids. By contrast all the 54 enzymes dedicated to this synthesis reported in the KEGG atlas for S. pombe were detected upon reannotation of S. pombe proteome (2 versus 54 against 278 versus 427, P<0.0001). This finding strongly suggests that species of the genus Pneumocystis are scavenging amino acids from their host's lung environment. Consequently, they would have no form able to live independently from another organism, and these parasites would be obligate in addition to being opportunistic. These findings have implications for the management of patients susceptible to P. jirovecii infection given that the only source of infection would be other humans.
Conflict of interest statement
Figures
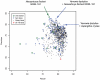
References
-
- Thomas CF, Jr, Limper AH. Current insights into the biology and pathogenesis of Pneumocystis pneumonia. Nat Rev Microbiol. 2007;5:298–308. - PubMed
-
- Thomas CF, Jr, Limper AH. Pneumocystis pneumonia. N Engl J Med. 2004;10:2487–2498. - PubMed
-
- Wakefield AE. Sattler F, Walzer PD, editors. Re-examination of epidemiological concepts. Bailliere's Clinical Infectious Diseases. 1995. pp. 431–448. In: Pneumocystis carinii.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

