Molecular basis of Wnt activation via the DIX domain protein Ccd1
- PMID: 21189423
- PMCID: PMC3048742
- DOI: 10.1074/jbc.M110.186742
Molecular basis of Wnt activation via the DIX domain protein Ccd1
Abstract
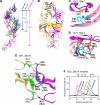
The Wnt signaling plays pivotal roles in embryogenesis and cancer, and the three DIX domain-containing proteins, Dvl, Axin, and Ccd1, play distinct roles in the initiation and regulation of canonical Wnt signaling. Overexpressed Dvl has a tendency to form large polymers in a cytoplasmic punctate pattern, whereas the biologically active Dvl in fact forms low molecular weight oligomers. The molecular basis for how the polymeric sizes of Dvl proteins are controlled upon Wnt signaling remains unclear. Here we show that Ccd1 up-regulates canonical Wnt signaling via acting synergistically with Dvl. We determined the crystal structures of wild type Ccd1-DIX and mutant Dvl1-DIX(Y17D), which pack into "head-to-tail" helical filaments. Structural analyses reveal two sites crucial for intra-filament homo- and hetero-interaction and a third site for inter-filament homo-assembly. Systematic mutagenesis studies identified critical residues from all three sites required for Dvl homo-oligomerization, puncta formation, and stimulation of Wnt signaling. Remarkably, Ccd1 forms a hetero-complex with Dvl through the "head" of Dvl-DIX and the "tail" of Ccd1-DIX, depolymerizes Dvl homo-assembly, and thereby controls the size of Dvl polymer. These data together suggest a molecular mechanism for Ccd1-mediated Wnt activation in that Ccd1 converts latent polymeric Dvl to a biologically active oligomer(s).
Figures
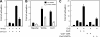





References
-
- Logan C. Y., Nusse R. (2004) Annu. Rev. Cell Dev. Biol. 20, 781–810 - PubMed
-
- Clevers H. (2006) Cell 127, 469–480 - PubMed
-
- Klaus A., Birchmeier W. (2008) Nat. Rev. Cancer 8, 387–398 - PubMed
-
- Cadigan K. M., Nusse R. (1997) Genes Dev. 11, 3286–3305 - PubMed
-
- Schwarz-Romond T., Fiedler M., Shibata N., Butler P. J., Kikuchi A., Higuchi Y., Bienz M. (2007) Nat. Struct. Mol. Biol. 14, 484–492 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

