Selective autophagy mediated by autophagic adapter proteins
- PMID: 21189453
- PMCID: PMC3060413
- DOI: 10.4161/auto.7.3.14487
Selective autophagy mediated by autophagic adapter proteins
Abstract
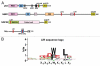
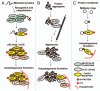
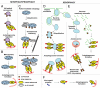
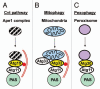
Mounting evidence suggests that autophagy is a more selective process than originally anticipated. The discovery and characterization of autophagic adapters, like p62 and NBR1, has provided mechanistic insight into this process. p62 and NBR1 are both selectively degraded by autophagy and able to act as cargo receptors for degradation of ubiquitinated substrates. A direct interaction between these autophagic adapters and the autophagosomal marker protein LC3, mediated by a so-called LIR (LC3-interacting region) motif, their inherent ability to polymerize or aggregate as well as their ability to specifically recognize substrates are required for efficient selective autophagy. These three required features of autophagic cargo receptors are evolutionarily conserved and also employed in the yeast cytoplasm-to-vacuole targeting (Cvt) pathway and in the degradation of P granules in C. elegans. Here, we review the mechanistic basis of selective autophagy in mammalian cells discussing the degradation of misfolded proteins, p62 bodies, aggresomes, mitochondria and invading bacteria. The emerging picture of selective autophagy affecting the regulation of cell signaling with consequences for oxidative stress responses, tumorigenesis and innate immunity is also addressed.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
