Mining electron density for functionally relevant protein polysterism in crystal structures
- PMID: 21190057
- PMCID: PMC3092063
- DOI: 10.1007/s00018-010-0611-4
Mining electron density for functionally relevant protein polysterism in crystal structures
Abstract
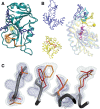
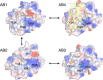
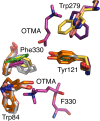
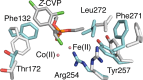
This review focuses on conceptual and methodological advances in our understanding and characterization of the conformational heterogeneity of proteins. Focusing on X-ray crystallography, we describe how polysterism, the interconversion of pre-existing conformational substates, has traditionally been analyzed by comparing independent crystal structures or multiple chains within a single crystal asymmetric unit. In contrast, recent studies have focused on mining electron density maps to reveal previously 'hidden' minor conformational substates. Functional tests of the importance of minor states suggest that evolutionary selection shapes the entire conformational landscape, including uniquely configured conformational substates, the relative distribution of these substates, and the speed at which the protein can interconvert between them. An increased focus on polysterism may shape the way protein structure and function is studied in the coming years.
Figures