Curvature recognition and force generation in phagocytosis
- PMID: 21190565
- PMCID: PMC3022777
- DOI: 10.1186/1741-7007-8-154
Curvature recognition and force generation in phagocytosis
Abstract
Background: The uptake of particles by actin-powered invagination of the plasma membrane is common to protozoa and to phagocytes involved in the immune response of higher organisms. The question addressed here is how a phagocyte may use geometric cues to optimize force generation for the uptake of a particle. We survey mechanisms that enable a phagocyte to remodel actin organization in response to particles of complex shape.
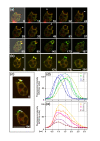
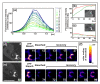
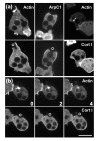
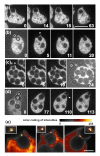
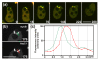
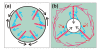
Results: Using particles that consist of two lobes separated by a neck, we found that Dictyostelium cells transmit signals concerning the curvature of a surface to the actin system underlying the plasma membrane. Force applied to a concave region can divide a particle in two, allowing engulfment of the portion first encountered. The phagosome membrane that is bent around the concave region is marked by a protein containing an inverse Bin-Amphiphysin-Rvs (I-BAR) domain in combination with an Src homology (SH3) domain, similar to mammalian insulin receptor tyrosine kinase substrate p53. Regulatory proteins enable the phagocyte to switch activities within seconds in response to particle shape. Ras, an inducer of actin polymerization, is activated along the cup surface. Coronin, which limits the lifetime of actin structures, is reversibly recruited to the cup, reflecting a program of actin depolymerization. The various forms of myosin-I are candidate motor proteins for force generation in particle uptake, whereas myosin-II is engaged only in retracting a phagocytic cup after a switch to particle release. Thus, the constriction of a phagocytic cup differs from the contraction of a cleavage furrow in mitosis.
Conclusions: Phagocytes scan a particle surface for convex and concave regions. By modulating the spatiotemporal pattern of actin organization, they are capable of switching between different modes of interaction with a particle, either arresting at a concave region and applying force in an attempt to sever the particle there, or extending the cup along the particle surface to identify the very end of the object to be ingested. Our data illustrate the flexibility of regulatory mechanisms that are at the phagocyte's disposal in exploring an environment of irregular geometry.
Figures






References
-
- Herant M, Heinrich V, Dembo M. Mechanics of neutrophil phagocytosis: behavior of the cortical tension. J Cell Sci. 2005;118:1789–1797. - PubMed
-
- Swanson JA, Johnson MT, Beningo K, Post P, Mooseker M, Araki N. A contractile activity that closes phagosomes in macrophages. J Cell Sci. 1999;112:307–316. - PubMed
-
- Beningo KA, Wang YL. Fc-receptor-mediated phagocytosis is regulated by mechanical properties of the target. J Cell Sci. 2002;115:849–856. - PubMed
-
- Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, Brückert F, Cosson P. Phg1p is a nine-transmembrane protein superfamily member involved in Dictyostelium adhesion and phagocytosis. J Biol Chem. 2000;275:34287–34292. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

