West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion
- PMID: 21191014
- PMCID: PMC3067947
- DOI: 10.1128/JVI.02050-10
West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion
Abstract
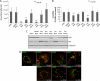
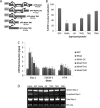
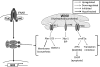
For intracellular survival it is imperative that viruses have the capacity to manipulate various cellular responses, including metabolic and biosynthetic pathways. The unfolded protein response (UPR) is induced by various external and internal stimuli, including the accumulation of misfolded proteins in the endoplasmic reticulum (ER). Our previous studies have indicated that the replication and assembly of the flavivirus West Nile virus strain Kunjin virus (WNV(KUN)) is intimately associated with the ER. Thus, we sought to determine whether the UPR was induced during WNV(KUN) infection. WNV(KUN) induces UPR signaling during replication, which is coordinated with peak replication. Interestingly, signaling is biased toward the ATF6/IRE-1 arm of the response, with high levels of Xbp-1 activation but negligible eukaryotic translation initiation factor 2α phosphorylation and downstream transcription. We show that the PERK-mediated response may partially regulate replication, since external UPR stimulation had a limiting effect on early replication events and cells deficient for PERK demonstrated increased replication and virus release. Significantly, we show that the WNV(KUN) hydrophobic nonstructural proteins NS4A and NS4B are potent inducers of the UPR, which displayed a high correlation in inhibiting Jak-STAT signaling in response to alpha interferon (IFN-α). Sequential removal of the transmembrane domains of NS4A showed that reducing hydrophobicity decreased UPR signaling and restored IFN-α-mediated activation. Overall, these results suggest that WNV(KUN) can stimulate the UPR to facilitate replication and that the induction of a general ER stress response, regulated by hydrophobic WNV(KUN) proteins, can potentiate the inhibition of the antiviral signaling pathway.
Figures


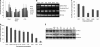
References
-
- Anthony, T. G., B. C. McGrath, D. R. Cavener, and R. C. Wek. 2003. The eIF2a kinase GCN2 is necessary for adaptation to dietary essential amino acid (EAA) deprivation in growing mice. FASEB J. 17:A811-A811.
-
- Bernales, S., F. R. Papa, and P. Walter. 2006. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22:487-508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

