Transcriptome analysis of grain-filling caryopses reveals involvement of multiple regulatory pathways in chalky grain formation in rice
- PMID: 21192807
- PMCID: PMC3023816
- DOI: 10.1186/1471-2164-11-730
Transcriptome analysis of grain-filling caryopses reveals involvement of multiple regulatory pathways in chalky grain formation in rice
Abstract
Background: Grain endosperm chalkiness of rice is a varietal characteristic that negatively affects not only the appearance and milling properties but also the cooking texture and palatability of cooked rice. However, grain chalkiness is a complex quantitative genetic trait and the molecular mechanisms underlying its formation are poorly understood.
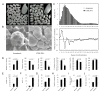
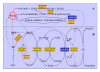
Results: A near-isogenic line CSSL50-1 with high chalkiness was compared with its normal parental line Asominori for grain endosperm chalkiness. Physico-biochemical analyses of ripened grains showed that, compared with Asominori, CSSL50-1 contains higher levels of amylose and 8 DP (degree of polymerization) short-chain amylopectin, but lower medium length 12 DP amylopectin. Transcriptome analysis of 15 DAF (day after flowering) caryopses of the isogenic lines identified 623 differential expressed genes (P < 0.01), among which 324 genes are up-regulated and 299 down-regulated. These genes were classified into 18 major categories, with 65.3% of them belong to six major functional groups: signal transduction, cell rescue/defense, transcription, protein degradation, carbohydrate metabolism and redox homeostasis. Detailed pathway dissection demonstrated that genes involved in sucrose and starch synthesis are up-regulated, whereas those involved in non-starch polysaccharides are down regulated. Several genes involved in oxidoreductive homeostasis were found to have higher expression levels in CSSL50-1 as well, suggesting potential roles of ROS in grain chalkiness formation.
Conclusion: Extensive gene expression changes were detected during rice grain chalkiness formation. Over half of these differentially expressed genes are implicated in several important categories of genes, including signal transduction, transcription, carbohydrate metabolism and redox homeostasis, suggesting that chalkiness formation involves multiple metabolic and regulatory pathways.
Figures





Similar articles
-
Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray.Plant Physiol. 2007 May;144(1):258-77. doi: 10.1104/pp.107.098665. Epub 2007 Mar 23. Plant Physiol. 2007. PMID: 17384160 Free PMC article.
-
Proteomic analysis of proteins related to rice grain chalkiness using iTRAQ and a novel comparison system based on a notched-belly mutant with white-belly.BMC Plant Biol. 2014 Jun 12;14:163. doi: 10.1186/1471-2229-14-163. BMC Plant Biol. 2014. PMID: 24924297 Free PMC article.
-
Dynamic formation and transcriptional regulation mediated by phytohormones during chalkiness formation in rice.BMC Plant Biol. 2021 Jun 30;21(1):308. doi: 10.1186/s12870-021-03109-z. BMC Plant Biol. 2021. PMID: 34193032 Free PMC article.
-
[Current status and strategies for improvement of rice grain chalkiness].Yi Chuan. 2009 Jun;31(6):563-72. doi: 10.3724/sp.j.1005.2009.00563. Yi Chuan. 2009. PMID: 19586854 Review. Chinese.
-
Proteomics and Post-Translational Modifications of Starch Biosynthesis-Related Proteins in Developing Seeds of Rice.Int J Mol Sci. 2021 May 31;22(11):5901. doi: 10.3390/ijms22115901. Int J Mol Sci. 2021. PMID: 34072759 Free PMC article. Review.
Cited by
-
Meta-QTL and haplo-pheno analysis reveal superior haplotype combinations associated with low grain chalkiness under high temperature in rice.Front Plant Sci. 2023 Mar 8;14:1133115. doi: 10.3389/fpls.2023.1133115. eCollection 2023. Front Plant Sci. 2023. PMID: 36968399 Free PMC article.
-
Agronomic and genetic analysis of Suweon 542, a rice floury mutant line suitable for dry milling.Rice (N Y). 2013 Dec 9;6(1):37. doi: 10.1186/1939-8433-6-37. Rice (N Y). 2013. PMID: 24321450 Free PMC article.
-
Biofortification of rice with lysine using endogenous histones.Plant Mol Biol. 2015 Feb;87(3):235-48. doi: 10.1007/s11103-014-0272-z. Epub 2014 Dec 17. Plant Mol Biol. 2015. PMID: 25512028 Free PMC article.
-
Complementary Proteome and Transcriptome Profiling in Developing Grains of a Notched-Belly Rice Mutant Reveals Key Pathways Involved in Chalkiness Formation.Plant Cell Physiol. 2017 Mar 1;58(3):560-573. doi: 10.1093/pcp/pcx001. Plant Cell Physiol. 2017. PMID: 28158863 Free PMC article.
-
Metabolomic analysis of pathways related to rice grain chalkiness by a notched-belly mutant with high occurrence of white-belly grains.BMC Plant Biol. 2017 Feb 7;17(1):39. doi: 10.1186/s12870-017-0985-7. BMC Plant Biol. 2017. PMID: 28166731 Free PMC article.
References
-
- Del-Rosario AR, Briones VP, Vidal AJ, Juliano BO. Composition and endosperm structure of developing and mature rice kernel. Cereal Chem. 1968;45:225–235.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

